Genetic Diversity and Population Structure of Boro Rice Landraces of Bangladesh
Received: 08-Jun-2022 / Manuscript No. rroa-22-66141 / Editor assigned: 10-Jun-2022 / PreQC No. rroa-22-66141 / Reviewed: 15-Jun-2022 / QC No. rroa-22-66141 / Revised: 18-Jun-2022 / Manuscript No. rroa-22-66141 / Published Date: 30-Jun-2022 DOI: 10.4172/2375-4338.1000305
Abstract
Molecular characterization, evaluation of genetic diversity and assessment of relationship by simple sequence repeat (SSR) markers of 48 Bangladeshi Boro rice landraces was performed. Among the total 58 SSR markers, 55 markers has been distributing over 12 rice chromosomes showed clear polymorphic band patterns, and they were selected for genetic relationship assessment. A total of 228 alleles were detected with an average of 4.15 alleles per locus. The average values of gene diversity and polymorphic information content (PIC) were 0.39 and 0.36, respectively. Primer RM206 had the highest PIC value (0.78) and the highest number of alleles (10).Therefore, RM206 was detected for the highest level of polymorphism and RM206 is supposed to be the best marker for characterizing the 48 Boro rice landraces. The genetic distance-based results in the unrooted neighbor-joining (NJ) tree revealed four (4) major clusters (I,II,III and IV)and a model-based population structure analysis generated two clusters (A and B). Both neighbors joining tree analysis and the population structure analysis method showed the tested landraces as highly diverse in structure. The two and three dimensional graphical views of Principal Coordinate Analysis (PCoA) revealed that the landraces Mi-Pajang, Gopal Beshi, Borail, Madhabsail, Boro (sunga) and Jala Boro were found far away and distributed around the centroid of the cluster. This rice collection and information gained in this study will be useful for future rice breeding program.
Keywords
Rice; Genetic diversity; Molecular characterization; Simple sequence repeat
Introduction
Rice has shaped the culture, diets and economic of thousands of millions of peoples. For more than half of the humanity “rice is life”. Bangladesh has abundant diversified rice landraces from time immemorial, since rice plays an important role in the livelihood, cultures and socio-economic aspects of the people, and is also the main cereal food in Bangladesh [1]. It has been cultivated in Asia since ancient times and for generations farmers have maintained thousands of different landraces [2]. Now, 90% of world rice is produced in Asia on an area of almost 150 million hectares. Rice accounts for 50% of agricultural income in Asia and supplies almost 80% of the region’s nutrition. In Bangladesh rice engages more than 70% of the rural population and is central to agriculture and the national economy [3].Due to great significance and intimate association of rice in food security and local ways of life and culture, Asian farmers have selected and maintained a vast array of over thousands of years. Scientists estimate that more than 1,40,000 rice varieties have been developed/selected/isolated in Asia [4]. More than 1,32,000 rice accessions and wild relatives can be found in the world’s largest Genebank for rice at IRRI (International Rice Research Institute) located in the Philippines (https://www.irri. org/international-rice-genebank). Until now, Bangladesh Rice Research Institute (BRRI) has collected and preserved more than 8,000 varieties/ landraces/cultivars/advanced lines/wild types from indigenous and exotic sources [4]. After establishment of BRRI, characterization or DNA fingerprinting has been done only for a small number of local landraces. Many countries in the world have characterized their indigenous different crop landraces at both molecular and phenotypic level. This has been done for keeping their crop identity and for searching new genes for further crop improvement. But information about the genetic diversity of local landraces as well as Boro rice is very limited. The needs for varietal improvement for such situations are very important.
Molecular markers are the powerful tools to detect genetic variation and genetic relationship within and among species. DNA markers are used for unmasking new genes and for the improvement of crop varieties [5]. The use of DNA markers has been suggested for precise and reliable characterization and discrimination of rice genotypes [6]. For genetic variability assessment, DNA markers are extensively used because they are not affected by environmental factors. Microsatellites (SSRs) are the marker of choice because of their advantages over other markers. These markers are polymorphic, abundant in eukaryotic organisms, and well distributed throughout the genome [7,8]. The SSRs are most suitable for rice because of their reproducibility, multiallelic nature, hypervariablility, codominant inheritance, relative abundance, and genome-wide coverage [9]. In addition, SSRs often have flanking regions that are highly conserved in related species, which allows the use of the same primer pairs in related genomes [10,11]. The SSR markers are particularly suitable for evaluating genetic diversity and relationships among plant species, populations, or individuals [12, 13]; studying rice landraces for either conservation or utilization [14]; marker-assisted selection breeding [15,16]; cultivar identification; and hybrid purity analysis and gene mapping studies [17,18,19,20]. Selection of parents based on genetic divergence using SSR markers has been successfully utilized in multiple crop species [21, 6].
Assessment of genetic diversity is very important in plant breeding or in biotechnology, if selection is the basis of improvement. For the assessment or analysis of genetic diversity, molecular markers are superior to morphological, pedigree, heterosis and biochemical data [22]. Genetic diversity is generally measured by genetic distance or genetic similarity, which imply that there are either differences or similarities at the genetic level of the plant [23]. Molecular marker based Genetic Diversity Analysis (MMGDA) also has potential for assessing changes in genetic diversity over time and space [24]. As the variation among the genotypes comes from the variations in DNA sequence, therefore, variations in DNA sequence are the basis of genetic diversity analysis [25]. Though, rice genome sequence is a valuable [26], most researchers are trying to identify particular segment of DNA or gene in a definite chromosome [25]. Molecular markers are the molecules that can trace a required gene in observed genotypes [27].
Information on the genetic diversity within and among closely related crop varieties is essential for a rational use of genetic resources and is of fundamental interest to plant breeders. It contributes to monitor landraces and can also be used to predict potential genetic gains [28]. Information regarding genetic variability at the molecular level could be used to help identify and develop genetically unique landraces that complements existing cultivars [29]. The objectives of this research were to assess the genetic variation and diversity of 48 Boro rice landraces, determine the genetic relationship among these landraces for breeding purposes and characterize these boro rice landraces.
Materials and Methods
Rice materials
In total, 48 Boro rice landrces of Bangladesh were studied (Table 1). Five gram seed from each of the entry was germinated and then sown in seedbed for growth and subsequent DNA extraction.
| Sl. No | Landraces | Acc. no. | Location of collection | Sl. No | Landraces | Acc. no. | Location of collection |
|---|---|---|---|---|---|---|---|
| G 1 | Mi-Pajang | 149 | Tangail | G 25 | Boro Dhan | 1808 | Kishorganj |
| G 2 | Dholi Boro | 180 | Tangail | G 26 | Boro Jagli | 1809 | Kishorganj |
| G 3 | Kumri Boro | 257 | Mymensingh | G 27 | Jagli | 1810 | Kishorganj |
| G 4 | Bairagi Sail | 261 | Mymensingh | G 28 | Deshi Boro | 1815 | Kishorganj |
| G 5 | Tepi Khorch | 931 | Sylhet | G 29 | Boro Dhan | 1816 | Kishorganj |
| G 6 | Pankaich | 937 | Sylhet | G 30 | Boro (Sunga) | 1861 | Dinajpur |
| G 7 | Boro Deshi | 938 | Sylhet | G 31 | Jala Boro | 1866 | Dinajpur |
| G 8 | Gopal Beshi | 939 | Sylhet | G 32 | Kali Boro 2/2 | 2189 | Gazipur |
| G 9 | Borail | 940 | Sylhet | G 33 | Kali Boro 4/1 | 2190 | Gazipur |
| G 10 | Boro 6/2 | 2206 | Gazipur | G 34 | Kali Boro 26 | 2191 | Gazipur |
| G 11 | Kali Boro | 1049 | Khulna | G 35 | Kali Boro 41/1 | 2192 | Gazipur |
| G 12 | Sonar Geye | 1050 | Khulna | G 36 | Kali Boro 48/1 | 2193 | Gazipur |
| G 13 | Joya Boro | 1051 | Khulna | G 37 | Kali Boro 80/3 | 2194 | Gazipur |
| G 14 | Amboro2 (Golden) | 1473 | Dhaka | G 38 | Kali Boro 80/5 | 2195 | Gazipur |
| G 15 | Batti Boro | 1477 | Dhaka | G 39 | Kali Boro 109/4 | 2196 | Gazipur |
| G 16 | Madhabsail | 1651 | Dhaka | G 40 | Kali Boro 138/2 | 2197 | Gazipur |
| G 17 | Jagli | 1704 | Faridpur | G 41 | Kali Boro 139/2 | 2198 | Gazipur |
| G 18 | Jagli | 1705 | Faridpur | G 42 | Kali Boro 200 | 2199 | Gazipur |
| G 19 | Local Boro | 1753 | Khulna | G 43 | Kali Boro 208 | 2200 | Gazipur |
| G 20 | Saita | 1794 | Kishorganj | G 44 | Kali Boro 259 | 2201 | Gazipur |
| G 21 | Dud Saita | 1795 | Kishorganj | G 45 | Kali Boro 266 | 2202 | Gazipur |
| G 22 | Bogra Boro | 1804 | Kishorganj | G 46 | Kali Boro 576 | 2203 | Gazipur |
| G 23 | Deshi Boro | 1805 | Kishorganj | G 47 | Kali Boro 600 | 2204 | Gazipur |
| G 24 | Jagli (DeshiBoro) | 1806 | Kishorganj | G 48 | Kali Boro 704 | 2205 | Gazipur |
Table 1: List of 48 Boro rice landraces.
SSR markers
A total of 55 SSR markers (Table 2) were found polymorphic and used for molecular characterization and diversity analysis.
Genomic DNA extraction and amplification
Total genomic DNA was extracted from young leaves of threeweek- old plants following the quick DNA extraction protocol of Ferdous et al. [30]. PCR analysis was performed in 10μl reaction sample containing 3μl of DNA template, 4.5 μl of GoTaq G2 Green Master Mix (Promega, USA), 1.5 μl of Nuclease-Free Water, 0.5 μl each of 10 μM forward and reverse primers using a GeneAtlas G (Astec, Japan) 96-well thermal cycler. The mixture was overlaid with 10 μl of mineral oil to prevent evaporation. After initial denaturation for five minutes at 94°C, each cycle comprised 30 sec denaturation at 95°C, 30 sec annealing at 55°C, and 25 sec extension with a final extension for 5 min at 72°C at the end of 32 cycles. The PCR products were analyzed by electrophoresis on 8% polyacrylamide gel with a 1 Kb plus DNA ladder (Thermo Scientific, USA) using mini vertica lpolyacrylamide gels for high throughput manual genotyping (CBS Scientific Co. Inc., CA, USA). 2.5 μl of amplification products were resolved by running gel in 0.5X TBE buffer for 1.5-2.5 hrs depending upon the allele size at around 100volts and 500 mA current. The gels were stained in 5 μl SYBR Safe DNA gel stain (10,000X concentration in DMSO, USA) with 200 ml 0.5X TBE buffer for 15 min and exposed to UV light using a molecular imager gel documentation unit (XR System, Uvitec Cambridge, France) for visualization. Microsatellite or simple sequence repeat (SSR) markers were used for molecular analysis [31,32]. We used fifty-five well-distributed SSRs for the diversity analysis; position (cM), repeat motifs, and chromosomal positions for the SSR markers can be found in the rice genome database (Gramene Portals, 2017). Most of these markers were obtained from a panel of fifty standard SSR markers, which has been proposed by CGIAR (Consultative Group for International Agricultural Research), for rice diversity analysis [33,34].
Data analysis
The band-size for each of the markers was scored using the AlphaEaseFC 4.0 software. Using PowerMarker version 3.25 [35], summary statistics included the following: the number of alleles, the major allele size and its frequency, gene diversity, and the polymorphism information content (PIC) value. For the unrooted phylogenic tree, the genetic distance was calculated using MEGA 6 based on Nei’s unbiased pairwise [36,37]. Binary form for allele frequency was prepared using PowerMarker software and used for dendrogram construction by NTSYS-pc software [38]. The unweighted pair grouping method using arithmetic average (UPGMA) was used to determine a similarity matrix following the Dice coefficient with the SAHN subprogram. Population STRUCTURE for landraces was determined using STRUCTURE, (version 2.3.4) [39, 40]. The number of clusters (K) investigated, in this study, ranged from one to fifteen, with five replications for analysis of each K value. The model following admixture and correlated allele frequency with a 5,000 burn period and a run length of 50,000 were used for conducting model-based structure analysis. Output of analysis was collected using the STRUCTURE harvester [41] and identified 4 as the best K value based on the LnP(D) and Evanno’s ΔK [42]. Principal components analysis (PCA) analysis was conducted also using the NTSYS-pc software.
Results and Discussion
SSR marker-based diversity and molecular characterization
All the 48Boro rice landraces distributed overall 12 chromosomes of rice were genotyped with 55 simple sequence repeat (SSR) markers. All 55 markers exhibiting polymorphism.
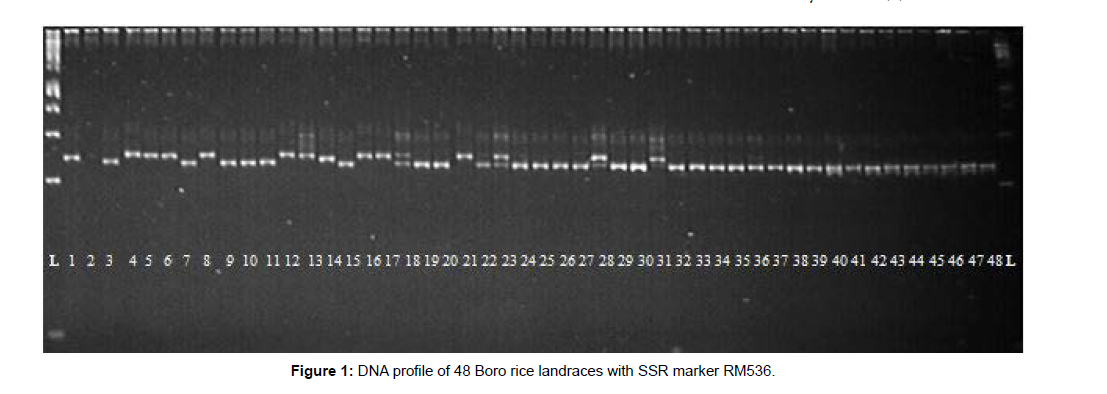
A total of 228 alleles were identified at 55 SSR markers over 48 Boro landraces (Table 2). RM484 (296bp) produced the maximum amplicon size and RM289 (88bp) was the minimum. In the case of RM472 (286- 306bp), a maximum range of band sizes was found and succeeded by RM484 (289-296bp) and RM591 (247-279 bp), respectively. The average number of alleles per locus was 4.15, ranged from 2 (RM133, RM145, RM212, RM316, RM320, RM338, RM411, RM452, RM455 and RM484) to 10 (RM206). The polymorphism information content (PIC) for the SSR loci ranged from 0.04 (RM455) to 0.78 (RM206) with an average of 0.36. Marker RM206 had the highest PIC value (0.78) and the highest number of alleles (10). SSRs which have higher PIC values have a higher number of alleles. Lower PIC value shows that the landraces under study are closely allied, whereas the higher value of PIC stipulates the higher array of materials which is the utmost need for parents selection for hybridization as well as for the new improved variety development, accordingly. Therefore, RM206 was detected as the highest level of polymorphism and RM206 is supposed to be the best marker for characterizing the 48 Boro rice landraces. The gene diversity ranged from 0.04 to 0.81, with an average of 0.39.The frequency of the most common allele at each locus ranged from 29.17% (RM206) to 97.92% (RM455). On average, 73.83% of the 48 rice landraces shared a common major allele at any given locus. The DNA profiles of 48 Boro rice landraces with RM536 are shown in (Figure 1).
| Sl. No. | Marker Name | Chro. No. |
Forward primer (5′–3′) | Reverse primer (5′–3′) | Annealing Temp. |
|---|---|---|---|---|---|
| 1 | RM1 | 1 | GCGAAAACACAATGCAAAAA | GCGTTGGTTGGACCTGAC | 55 |
| 2 | RM5 | 1 | TGCAACTTCTAGCTGCTCGA | GCATCCGATCTTGATGGG | 55 |
| 3 | RM212 | 1 | CCACTTTCAGCTACTACCAG | CACCCATTTG | 55 |
| 4 | RM472 | 1 | CCATGGCCTGAGAGAGAGAG | AGCTAAATGGCCATACGGTG | 55 |
| 5 | RM283 | 1 | GTCTACATGTACCCTTGTTGGG | CGGCATGAGAGTCTGTGATG | 55 |
| 6 | RM495 | 2 | AATCCAAGGTGCAGAGATGG | CAACGATGACGAACACAACC | 55 |
| 7 | RM307 | 2 | GTACTACCGACCTACCGTTCAC | CTGCTATGCATGAACTGCTC | 55 |
| 8 | RM145 | 2 | CCGGTAGGCGCCCTGCAGTTTC | CAAGGACCCCATCCTCGGCGTC | 56 |
| 9 | RM208 | 2 | TCTGCAAGCCTTGTCTGATG | TAAGTCGATCATTGTGTGGACC | 55 |
| 10 | RM213 | 2 | ATCTGTTTGCAGGGGACAAG | AGGTCTAGACGATGTCGTGA | 55 |
| 11 | RM262 | 2 | CATTCCGTCTCGGCTCAACT | CAGAGCAAGGTGGCTTGC | 55 |
| 12 | RM263 | 2 | GCGCTGGTGGAAAATGAG | GGCATCCCTCTTTGATTCCTC | 55 |
| 13 | RM452 | 2 | CTGATCGAGAGCGTTAAGGG | GGGATCAAACCACGTTTCTG | 55 |
| 14 | RM207 | 2 | CCATTCGTGAGAAGATCTGA | CACCTCATCCTCGTAACGCC | 55 |
| 15 | RM411 | 3 | ACACCAACTCTTGCCTGCAT | TGAAGCAAAAACATGGCTAGG | 55 |
| 16 | RM520 | 3 | AGGAGCAAGAAAAGTTCCCC | GCCAATGTGTGACGCAATAG | 55 |
| 17 | RM3646 | 3 | ACTAGAGCACCCTCGCTGAG | CTCAGCCACCCCATCAAC | 55 |
| 18 | RM338 | 3 | CACAGGAGCAGGAGAAGAGC | GGCAAACCGATCACTCAGTC | 55 |
| 19 | RM16 | 3 | CGCTAGGGCAGCATCTAAA | AACACAGCAGGTACGCGC | 55 |
| 20 | RM252 | 4 | TTCGCTGACGTGATAGGTTG | ATGACTTGATCCCGAGAACG | 55 |
| 21 | RM273 | 4 | GAAGCCGTCGTGAAGTTACC | GTTTCCTACCTGATCGCGAC | 55 |
| 22 | RM303 | 4 | GCATGGCCAAATATTAAAGG | GGTTGGAAATAGAAGTTCGGT | 55 |
| 23 | RM334 | 5 | GTTCAGTGTTCAGTGCCACC | GACTTTGATCTTTGGTGGACG | 55 |
| 24 | RM26 | 5 | GAGTCGACGAGCGGCAGA | CTGCGAGCGACGGTAACA | 55 |
| 25 | RM289 | 5 | TTCCATGGCACACAAGCC | CTGTGCACGAACTTCCAAAG | 55 |
| 26 | RM413 | 5 | GGCGATTCTTGGATGAAGAG | TCCCCACCAATCTTGTCTTC | 55 |
| 27 | RM190 | 6 | CTTTGTCTATCTCAAGACAC | TTGCAGATGTTCTTCCTGATG | 55 |
| 28 | RM170 | 6 | TCGCGCTTCTTCCTCGTCGACG | CCCGCTTGCAGAGGAAGCAGCC | 55 |
| 29 | RM125 | 6 | ATCAGCAGCCATGGCAGCGACC | AGGGGATCATGTGCCGAAGGCC | 55 |
| 30 | RM133 | 6 | TTGGATTGTTTTGCTGGCTCGC | GGAACACGGGGTCGGAAGCGAC | 61 |
| 31 | RM253 | 6 | TCCTTCAAGAGTGCAAAACC | GCATTGTCATGTCGAAGCC | 55 |
| 32 | RM510 | 7 | AACCGGATTAGTTTCTCGCC | TGAGGACGACGAGCAGATTC | 55 |
| 33 | RM320 | 7 | CAACGTGATCGAGGATAGATC | GGATTTGCTTACCACAGCTC | 55 |
| 34 | RM455 | 7 | AACAACCCACCACCTGTCTC | AGAAGGAAAAGGGCTCGATC | 55 |
| 35 | RM223 | 8 | GAGTGAGCTTGGGCTGAAAC | GAAGGCAAGTCTTGGCACTG | 55 |
| 36 | RM342 | 8 | CCATCCTCCTACTTCAATGAAG | ACTATGCAGTGGTGTCACCC | 55 |
| 37 | RM515 | 8 | TAGGACGACCAAAGGGTGAG | TGGCCTGCTCTCTCTCTCTC | 55 |
| 38 | RM447 | 8 | CCCTTGTGCTGTCTCCTCTC | ACGGGCTTCTTCTCCTTCTC | 55 |
| 39 | RM201 | 9 | CTCGTTTATTACCTACAGTACC | CTACCTCCTTTCTAGACCGATA | 55 |
| 40 | RM205 | 9 | CTGGTTCTGTATGGGAGCAG | CTGGCCCTTCACGTTTCAGTG | 55 |
| 41 | RM316 | 9 | CTAGTTGGGCATACGATGGC | ACGCTTATATGTTACGTCAAC | 55 |
| 42 | RM228 | 10 | CTGGCCATTAGTCCTTGG | GCTTGCGGCTCTGCTTAC | 55 |
| 43 | RM304 | 10 | TCAAACCGGCACATATAAGAC | GATAGGGAGCTGAAGGAGATG | 55 |
| 44 | RM591 | 10 | CTAGCTAGCTGGCACCAGTG | TGGAGTCCGTGTTGTAGTCG | 55 |
| 45 | RM484 | 10 | TCTCCCTCCTCACCATTGTC | TGCTGCCCTCTCTCTCTCTC | 55 |
| 46 | RM536 | 11 | TCTCTCCTCTTGTTTGGCTC | ACACACCAACACGACCACAC | 55 |
| 47 | RM202 | 11 | CAGATTGGAGATGAAGTCCTCC | CCAGCAAGCATGTCAATGTA | 55 |
| 48 | RM206 | 11 | CCCATGCGTTTAACTATTCT | CGTTCCATCGATCCGTATGG | 55 |
| 49 | RM209 | 11 | ATATGAGTTGCTGTCGTGCG | CAACTTGCATCCTCCCCTCC | 55 |
| 50 | RM144 | 11 | TGCCCTGGCGCAAATTTGATCC | GCTAGAGGAGATCAGATGGTAGGCATG | 55 |
| 51 | RM224 | 11 | ATCGATCGATCTTCACGAGG | TGCTATAAAAGGCATTCGGG | 55 |
| 52 | RM19 | 12 | CAAAAACAGAGCAGATGAC | CTCAAGATGGACGCCAAGA | 55 |
| 53 | RM277 | 12 | CGGTCAAATCATCACCTGAC | CAAGGCTTGCAAGGGAAG | 55 |
| 54 | RM1337 | 12 | GTGCAATGCTGAGGAGTATC | CTGAGAATCTGGAGTGCTTG | 55 |
| 55 | RM12 | 12 | TGCCCTGTTATTTTCTTCTCTC | GGTGATCCTTTCCCATTTCA | 55 |
Table 2: List of 55 SSR markers.
In recent past, molecular characterization, genetic diversity and population structure of Bangladeshi rice landraces have been studied by using molecular (SSR) markers [1, 36, 34, 43].
From our present study, the genetic diversity is similar to earlier studies [29], they identified 4.18 alleles per locus and an average PIC value of 0.488 among 21 rice landraces (Table 3). Also, the average PIC value 0.44 was observed among 43 Thai and 57 IRRI landraces of rice by Chakhonkaen et al. [45]. But, Ahmed et al. [1] found 350 alleles from similarly named rice landraces using 45 SSR markers and several alleles per locus ranged from 3 to 14 with an average of 7.8 which was higher than our present study. On the other hand, a lower genetic diversity was disclosed among 50 Bangladeshi red rice varieties were collected from different agro-climatic regions of Bangladesh having 3.24 alleles per locus with mean PIC value of 0.32, also reported by Islam et al. [34]. Also, a marginally lower genetic diversity was disclosed among 28 restorer lines of hybrid rice with an average of 2.67 alleles per locus and an average PIC value of 0.29 [46].
| SL. No. | Marker | Chromosome No. |
Position (cM) |
Motif* | Allele No. |
Unique allele |
Size range (bp) |
Size (bp) | Frequency (%) | Gene diversity | PIC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | RM1 | 1 | 29.7 | (GA)26 | 4 | - | 80-112 | 112 | 54.17 | 0.36 | 0.34 |
| 2 | RM5 | 1 | 94.9 | GA)14 | 4 | - | 111-127 | 111 | 43.75 | 0.63 | 0.56 |
| 3 | RM16 | 3 | 131.5 | (TCG)5(GA)16 | 4 | 1 | 184-240 | 184 | 85.42 | 0.26 | 0.25 |
| 4 | RM12 | 12 | 109.1 | (GA)21 | 6 | 2 | 150-215 | 150 | 72.92 | 0.45 | 0.43 |
| 5 | RM19 | 12 | 20.9 | (ATC)10 | 7 | 4 | 204-239 | 239 | 77.08 | 0.39 | 0.38 |
| 6 | RM26 | 5 | 118.8 | (GA)15 | 3 | - | 105-119 | 119 | 89.58 | 0.19 | 0.18 |
| 7 | RM133 | 6 | 0 | (CT)8 | 2 | - | 225-234 | 234 | 93.75 | 0.12 | 0.11 |
| 8 | RM144 | 11 | 123.2 | (ATT)11 | 5 | 1 | 220-247 | 247 | 54.17 | 0.63 | 0.58 |
| 9 | RM145 | 2 | 49.8 | - | 2 | - | 181-192 | 181 | 95.83 | 0.08 | 0.08 |
| 10 | RM170 | 6 | 2.2-7.4 | (CCT7) | 5 | - | 98-127 | 127 | 60.42 | 0.58 | 0.54 |
| 11 | RM190 | 6 | 7.4 | (CT)11 | 3 | - | 107-127 | 127 | 85.42 | 0.26 | 0.24 |
| 12 | RM201 | 9 | 81.2 | (CT)17 | 5 | 1 | 161-188 | 161 | 68.75 | 0.50 | 0.47 |
| 13 | RM202 | 11 | 54 | (CT)30 | 4 | - | 165-186 | 186 | 64.58 | 0.54 | 0.50 |
| 14 | RM205 | 9 | 114.7 | (CT)25 | 5 | 1 | 117-156 | 117 | 64.58 | 0.54 | 0.51 |
| 15 | RM206 | 11 | 102.9 | (CT)21 | 10 | 4 | 135-198 | 135 | 29.17 | 0.81 | 0.78 |
| 16 | RM207 | 2 | 191.2 | (CT)25 | 4 | 1 | 72-148 | 148 | 79.17 | 0.36 | 0.33 |
| 17 | RM208 | 2 | 186.4 | (CT)17 | 3 | - | 170-187 | 170 | 72.92 | 0.43 | 0.38 |
| 18 | RM209 | 11 | 73.9 | (CT)18 | 7 | 2 | 125-170 | 125 | 60.42 | 0.60 | 0.58 |
| 19 | RM212 | 1 | 148.7 | (CT)24 | 2 | - | 125-132 | 132 | 87.50 | 0.22 | 0.19 |
| 20 | RM213 | 2 | 186.4 | (CT)17 | 3 | - | 143-166 | 143 | 85.42 | 0.26 | 0.24 |
| 21 | RM223 | 8 | 80.5 | (CT)25 | 4 | 1 | 144-172 | 172 | 72.92 | 0.44 | 0.40 |
| 22 | RM224 | 11 | 120.1 | (AAG)8(AG)13 | 6 | 2 | 138-172 | 172 | 56.25 | 0.60 | 0.55 |
| 23 | RM228 | 10 | 130.3 | (CA)6(GA)36 | 4 | - | 108-145 | 145 | 77.08 | 0.38 | 0.36 |
| 24 | RM252 | 4 | 99 | (CT)19 | 5 | 1 | 206-230 | 206 | 75.00 | 0.42 | 0.39 |
| 25 | RM253 | 6 | 37 | (GA)25 | 4 | 1 | 125-160 | 160 | 72.92 | 0.44 | 0.41 |
| 26 | RM262 | 2 | 103.3 | (CT)16 | 5 | 2 | 145-167 | 145 | 70.83 | 0.47 | 0.44 |
| 27 | RM263 | 2 | 127.5 | (CT)34 | 4 | - | 192-217 | 199 | 85.42 | 0.26 | 0.25 |
| 28 | RM273 | 4 | 94.4 | (GA)11 | 3 | - | 127-222 | 222 | 89.58 | 0.19 | 0.18 |
| 29 | RM277 | 12 | 57.2 | (GA)11 | 3 | 1 | 124-138 | 124 | 89.58 | 0.19 | 0.18 |
| 30 | RM283 | 1 | 31.4 | (GA)18 | 3 | - | 159-174 | 159 | 66.67 | 0.49 | 0.43 |
| 31 | RM289 | 5 | 56.7 | G11(GA)16 | 5 | - | 88-146 | 88 | 39.58 | 0.70 | 0.65 |
| 32 | RM304 | 10 | 73 | (GT)2(AT)10GT)33 | 4 | - | 157-174 | 157 | 45.83 | 0.63 | 0.57 |
| 33 | RM316 | 9 | 1.8 | (GT)8(TG)9(TTTG)4(TG)4 | 2 | - | 205-212 | 212 | 89.58 | 0.19 | 0.17 |
| 34 | RM320 | 7 | 62.4 | (AT)11GTAT (GT)13 |
2 | - | 225-233 | 233 | 95.83 | 0.08 | 0.08 |
| 35 | RM338 | 3 | 108.4 | (CTT)6 | 2 | - | 191-198 | 191 | 95.83 | 0.08 | 0.08 |
| 36 | RM342 | 8 | 78.4 | (CAT)12 | 5 | 3 | 130-169 | 130 | 81.25 | 0.32 | 0.30 |
| 37 | RM411 | 3 | 127.9 | (GTT)7 | 2 | - | 108-115 | 108 | 85.42 | 0.25 | 0.22 |
| 38 | RM413 | 5 | 26.7 | (AG)11 | 4 | - | 170-198 | 170 | 58.33 | 0.59 | 0.54 |
| 39 | RM447 | 8 | 124.6 | (CTT)8 | 5 | 2 | 106-145 | 106 | 83.33 | 0.30 | 0.28 |
| 40 | RM452 | 2 | 58.4 | (GTC)9 | 2 | - | 197-206 | 206 | 79.17 | 0.33 | 0.28 |
| 41 | RM455 | 7 | 65.7 | (TTCT)5 | 2 | 1 | 125-131 | 131 | 97.92 | 0.04 | 0.04 |
| 42 | RM484 | 10 | 97.3 | (AT)9 | 2 | - | 289-296 | 296 | 91.67 | 0.15 | 0.14 |
| 43 | RM495 | 2 | 2.8 | (CTG)7 | 3 | - | 159-173 | 159 | 83.33 | 0.29 | 0.26 |
| 44 | RM515 | 8 | 80.5 | (GA)11 | 6 | 1 | 211-252 | 211 | 72.92 | 0.45 | 0.43 |
| 45 | RM520 | 3 | 191.6 | (AG)10 | 3 | - | 244-264 | 244 | 70.83 | 0.45 | 0.40 |
| 46 | RM536 | 11 | 55.1 | (CT)16 | 3 | - | 236-249 | 249 | 72.92 | 0.41 | 0.36 |
| 47 | RM591 | 10 | 118.3 | (AC)10 | 5 | 1 | 247-279 | 247 | 62.50 | 0.56 | 0.53 |
| 48 | RM1337 | 12 | 57.2 | (GA)11 | 5 | - | 194-225 | 194 | 75.00 | 0.42 | 0.39 |
| 49 | RM3646 | 3 | 6.32 | (GA)14 | 3 | - | 145-158 | 145 | 85.42 | 0.26 | 0.24 |
| 50 | RM303 | 4 | 116.9 | [AC(AT)210]9 (GT)7(ATGT)6] |
8 | 1 | 107-198 | 198 | 54.17 | 0.67 | 0.64 |
| 51 | RM307 | 4 | 191.2 | (CT)25 | 7 | 2 | 125-264 | 125 | 77.08 | 0.40 | 0.38 |
| 52 | RM334 | 5 | 141.8 | (CTT)48 | 7 | 1 | 156-218 | 156 | 45.83 | 0.72 | 0.68 |
| 53 | RM472 | 1 | 171.6 | (GA)21 | 3 | - | 286-306 | 286 | 58.33 | 0.57 | 0.50 |
| 54 | RM125 | 7 | 24.8 | (GCT)8 | 4 | 1 | 127-152 | 127 | 85.42 | 0.26 | 0.25 |
| 55 | RM510 | 6 | 20.8 | (GA)15 | 3 | 1 | 113-127 | 127 | 91.67 | 0.16 | 0.15 |
| Max. | 10.00 | 296 | 97.92 | 0.81 | 0.78 | ||||||
| Min. | 2.00 | 88 | 29.17 | 0.04 | 0.04 | ||||||
| Total | 228.00 | 39 | 4060.42 | 21.35 | 19.82 | ||||||
| Mean | 4.15 | 73.83 | 0.39 | 0.36 |
Table 3: Number of alleles, allele size range, frequency, gene diversity and polymorphism information content (PIC) among 48 Boro rice landraces against 55 microsatellite markers.
Legend: 1. Mi-Pajang, 2. Dholi Boro, 3. Kumri Boro, 4. Bairagi Sail 5. Tepi Khorch 6. Pankaich, 7. BoroDeshi, 8. Gopal Beshi, 9. Borail, 10. Boro 6/2, 11. Kali Boro, 12. Sonar Geye, 13. Joya Boro, 14. Amboro2 (Golden), 15. Batti Boro, 16. Madhabsail, 17. Jagli, 18. Jagli, 19. Local Boro, 20. Saita, 21.Dud Saita, 22. Bogra Boro, 23. Deshi Boro, 24. Jagli (DeshiBoro), 25. Boro Dhan, 26.Boro Jagli, 27. Jagli, 28. Deshi Boro, 29. BoroDhan, 30.Boro (Sunga), 31. Jala Boro, 32. Kali Boro 2/2, 33. Kali Boro 4/1, 34. Kali Boro 26, 35. Kali Boro 41/1, 36. Kali Boro 48/1, 37. Kali Boro 80/3, 38. Kali Boro 80/5, 39. Kali Boro 109/4, 40. Kali Boro 138/2, 41. Kali Boro 139/2, 42. Kali Boro 200, 43. Kali Boro 208, 44. Kali Boro 259, 45. Kali Boro 266, 46. Kali Boro 576, 47. Kali Boro 600, 48. Kali Boro 704, L= 1Kb plus Ladder.
Unique Alleles
A total of 39 unique alleles were detected at 25 microsatellite loci. Nineteen (19) of the rice landraces had unique alleles for at least one microsatellite locus. Notably, seven (7) SSR markers—RM144 (chromosome 11, 220bp), RM205 (chromosome 9, 117bp), RM206 (chromosome 11, 152bp), RM277 (chromosome 12, 138bp), RM342 (chromosome 8, 152bp), RM515 (chromosome 8, 211bp) and RM307 (chromosome 4, 125bp) amplified specific alleles only in a slender grain rice landrace Jala Boro (Acc. No-1866). Similarly, six (6) SSR markers—RM19 (chromosome 12, 239bp), RM224 (chromosome 11, 138bp), RM262 (chromosome 2, 167bp), RM455 (chromosome 7, 125bp), RM125 (chromosome 7, 152bp) and RM510 (chromosome 6, 113bp) amplified specific alleles only in rice landrace Mi-Pajang (Acc. No-149). Also, Tepi Khorch (Acc. No-931) showed unique alleles in 4 markers and Bairagi Sail (Acc. No-261), Pankaich (Acc. No-937) and Joya Boro (Acc. No-1051) showed unique alleles in 3 markers (Table 4).
| SL. No. | Marker | Chromosome No. | Unique Allele (bp) | Landraces |
|---|---|---|---|---|
| 1 | RM16 | 3 | 184 | G 30 (Boro (sunga)) |
| 2 | RM12 | 12 | 183 | G 6(Pankaich) |
| 3 | RM12 | 12 | 208 | G 15(Batti Boro) |
| 4 | RM19 | 12 | 204 | G 9 (Borail) |
| 5 | RM19 | 12 | 216 | G 4 (Bairagi Sail) |
| 6 | RM19 | 12 | 227 | G 2 (Dholi Boro) |
| 7 | RM19 | 12 | 239 | G 1 (Mi-Pajang) |
| 8 | RM144 | 11 | 220 | G 31(Jala Boro) |
| 9 | RM201 | 9 | 188 | G 40 (Kali Boro 138/2) |
| 10 | RM205 | 9 | 117 | G 31 (Jala Boro) |
| 11 | RM206 | 11 | 152 | G 31 (Jala Boro) |
| 12 | RM206 | 11 | 171 | G 32 (Kali Boro 2/2) |
| 13 | RM206 | 11 | 192 | G 28 (Deshi Boro) |
| 14 | RM206 | 11 | 198 | G 7 (Boro Deshi) |
| 15 | RM207 | 2 | 148 | G 5 (Tepi Khorch) |
| 16 | RM209 | 11 | 134 | G 13 (Joya Boro) |
| 17 | RM209 | 11 | 176 | G 4 (Bairagi Sail) |
| 18 | RM223 | 8 | 172 | G 5 (Tepi Khorch) |
| 19 | RM224 | 11 | 138 | G 1 (Mi-Pajang) |
| 20 | RM224 | 11 | 143 | G 5 (Tepi Khorch) |
| 21 | RM252 | 4 | 230 | G 16 (Madhabsail) |
| 22 | RM253 | 6 | 160 | G 5 (Tepi Khorch) |
| 23 | RM262 | 2 | 161 | G 13 (Joya Boro) |
| 24 | RM262 | 2 | 167 | G 1 (Mi-Pajang) |
| 25 | RM277 | 12 | 138 | G 31 (Jala Boro) |
| 26 | RM342 | 8 | 146 | G 14 (Amboro 2 (golden)) |
| 27 | RM342 | 8 | 152 | G 31 (Jala Boro) |
| 28 | RM342 | 8 | 169 | G 6 (Pankaich) |
| 29 | RM447 | 8 | 121 | G 13 (Joya Boro) |
| 30 | RM447 | 8 | 145 | G 18 (Jagli) |
| 31 | RM455 | 7 | 125 | G 1 (Mi-Pajang) |
| 32 | RM515 | 8 | 211 | G 31 (Jala Boro) |
| 33 | RM591 | 10 | 266 | G 4 (Bairagi Sail) |
| 34 | RM303 | 4 | 107 | G 21 (Dud Saita) |
| 35 | RM307 | 4 | 125 | G 31 (Jala Boro) |
| 36 | RM307 | 4 | 147 | G 6 (Pankaich) |
| 37 | RM334 | 5 | 172 | G 12 (Sonar Geye) |
| 38 | RM125 | 7 | 152 | G 1 (Mi-Pajang) |
| 39 | RM510 | 6 | 113 | G 1 (Mi-Pajang) |
Table 4: List of identified unique alleles along with markers for 19 Boro rice landraces.
Unique alleles are precious as they June be effectively indicator of particular landraces and also for a breeding purpose. Generally, the higher number of unique alleles in landraces specifies as a sources of novel alleles. The unique alleles for molecular characterization were also detected by Siwach et al. [47], Das et al. [48] and Islam et al. [34]. Siwach et al. [47] identified seven SSR markers (RM1, RM21, RM38,RM170, RM210, RM226, and RM229) that showed unique alleles which could distinguish basmati rice varieties from non-basmati rice varieties. Also, Saini et al. [49] noticed 58 unique alleles among Basmati and non-Basmati rice varieties.
Genetic distance-based analysis
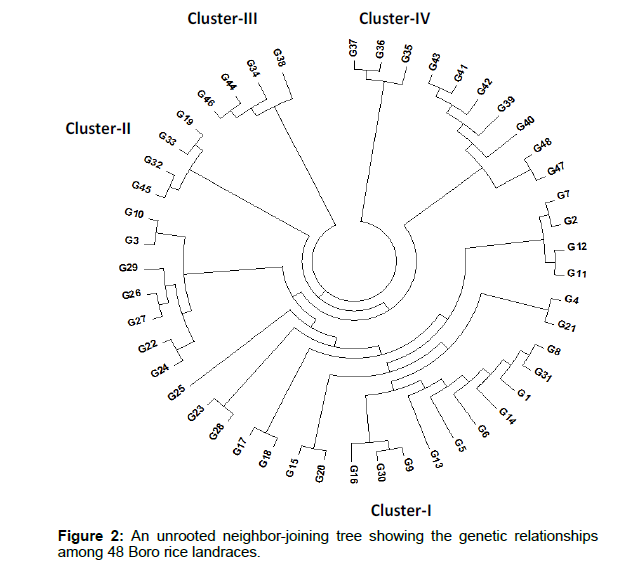
The genetic distance-based results seen in the unrooted neighborjoining (NJ) tree revealed that the 48 Boro rice landraces were grouped into four major clusters (Figure 2). The highest numbers of landraces (37) were found in cluster I followed by clusters II (4), III (4) and the lowest in cluster IV (3).
Genetic distance is useful for assessing the diversity between species and populations within a species. The highest number of germplasm (37) was found in cluster I. Again, cluster I was further divided into 2 sub clusters (A and B). Sub cluster-A, contains 30 landraces and Sub cluster-B consisted of 7 landraces. Cluster II contains 4 landraces (Local Boro, Kali Boro 2/2, Kali Boro 4/1 and Kali Boro266) and cluster III also contains 4 boro landraces (Kali Boro 26, Kali Boro 80/5, Kali Boro 259 and Kali Boro 576). The lowest number (3) of boro rice landraces was found in cluster IV (Kali Boro41/1, Kali Boro 48/1 and Kali Boro 80/3). In previous studies, Islam et al. [34] and Islam et al. [43] both found three (3) major clusters and few sub-clusters through the unrooted neighbor-joining (NJ) tree among 50 Bangladeshi red rice varieties and 113 aromatic rice landraces, respectively.
Population structure based on Model
Model-based program STRUCTURE 2.3.4 was used to find out the genetic relationship among individual Boro rice landraces. By the
admixture model, simulations were executed with K, range from 2
to 10 with 5 repetitions employing 48 landraces and the population
structure analysis declared the log-likelihood value (ΔK) maximized to
the highest value of at K= 2 (Figure 3), demonstrating a sharp peak
expressing the classification of entire landraces into two specific subgroups,
here denoted as Group A and Group B, respectively. To find
out the number of pure and admixed individuals, populations were
studied. The population Group A (red colour, Figure 4) and Group B
(green colour, Figure 4) representing 75% (36) and 25% (12) of Boro
landraces used in structure analysis, respectively. Overall, 11 (22.92%)
admixed landraces were found at K = 2. It June be noted that Group
A had 36 Boro rice landraces with 28 pure and 08 admixed landraces
and Group B had 12 boro rice landraces with 09 pure and 03 admixed
landraces. A similar population structure of two groups was observed
in previous research by Pathaichindachote et al. [50] in a collection of
167 Thai and exotic rice accessions in Thailand. Islam et al. [43] have
successfully classified three groups as indica aromatic rice landraces
in Bangladesh and again, Islam et al. [
Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Cross Ref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar,Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at, Google Scholar, Crossref Indexed at,Google Scholar, Crossref
Citation:
Khalequzzaman M, Islam MZ, Prince FRK, Rashid ESH, Siddique A (2022) Genetic Diversity and Population Structure of Boro Rice Landraces of Bangladesh. J Rice Res 10: 305. DOI: 10.4172/2375-4338.1000305 Copyright: © 2022 Khalequzzaman M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.References
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 3782
- [From(publication date): 0-2022 - Nov 18, 2025]
- Breakdown by view type
- HTML page views: 3270
- PDF downloads: 512