Genetic Divergence in Ethiopian Coriander (Coriandrum sativum l.) Accessions
Received: 19-Sep-2013 / Accepted Date: 24-Oct-2013 / Published Date: 26-Oct-2013 DOI: 10.4172/2329-8863.1000116
Abstract
Although Ethiopia is a center of diversity for many crops including Coriander little is known on the genetic divergence of this crop due to its negligence in the research program of the country in the past. The genetic divergence of 25 land races was assessed using principal component and cluster analysis based on 8 characters. The accessions were grouped into five clusters. Cluster I was the largest consisting 19 accessions. High inter cluster distance (47.42) was observed between cluster IV, cluster II and IV (47.33) and cluster I and IV (41.47) indicating the presence of substantial genetic diversity in genetic makeup of the accessions included in these clusters. Accessions 16 and 8 having positive values for principal component 1 and 2 were of considerable breeding interest because of their good combination for the studied yield related traits.
Keywords: Coriander accessions, Genetic divergence, Cluster analysis, Principal component analysis
402263Introduction
Coriander (Coriandrum sativum L.) is an annual spice herb that belongs to the family of Umbelliferae/Apiaceae. It is used as a spice, medicine and a raw material in food, beverage and pharmaceutical industries. Its green foliage, rich in vitamins and other minerals, is used in vegetables and salads while its seeds contain essential oils rich in linalool [1]. Although coriander is one of the several plant species for which Ethiopia is known as a center of origin and diversity [2], there is little information on its genetic divergence which in turn hinders the exploitation of the wealth of its diversity. The only work so far done on genetic divergence on Ethiopian coriander is that of Mengesha et al. [3], that focused on collections from different agroecological and geographical areas of the country. However, ecological and geographical diversifications are not the only causes of genetic divergence. For the changing of genetic material, genetic drift, natural variation and artificial selection also contribute to the genetic divergence [4] coriander accessions were diversified in different agroecologies of Ethiopia. Therefore, intensive collection focusing on the desired traits will benefit breeders by large for effective improvement in coriander [3]. Accordingly, target collection of the present coriander accessions was made from the potential growing areas of Arsi and Bale zones that were not well covered before and thus not well addressed by work of Mengesha et al. [3].
Genetic divergence is an essential prerequisite factor in any crop improvement programme to identify potential parents for hybridization and to obtain high yielding variety [5]. Therefore, having precise information and knowledge on the nature and degree of genetic divergence is helpful and fundamental to identify and organize the available genetic resources aiming at the production of promising cultivars [6]. For the selection of parents based on the extent of genetic divergence in different crop species multivariate methods have successfully utilized [7]. Target collection of coriander accession was made from the present study was undertaken with the following objectives: a) to assess and evaluate genetic diversity of coriander accessions, b) to identify characters which contribute at maximum to genetic diversity and c) to identify accessions for future use in breeding programs in coriander.
Materials and Methods
The experiment was conducted at Sinana Agricultural Research Center in 2012 which is located at an altitude of 2400 m.a.s.l. Sinana has a range of mean annual rainfall of 563-1018 mm with minimum and maximum temperature of 7.9 °C and 24.3°C, respectively. The soil type is dark-brown with slightly acidic reaction [8].
Twenty five coriander accessions collected from Arsi and Bale potential growing areas were sown in RCBD with three replications on a plot of 2 meter length with spacing of 15 and 30 cm between plants and rows, respectively. The experiment was conducted under rain-fed condition. Three times hoeing and weeding were carried out without the application of chemicals and fertilizers. Five plants were randomly selected for the measurement of the characters. A total of 8 characters were recorded according to the descriptors of International Plant Genetic Resource Institute (IPGRI) as given by Diederichsen [2]. These are number of basal leaves, length of basal leave, length of the longest basal leaves, habitus of the basal leaves, blade shape of the upper stem leaves, blade shape of the longest basal leaves, foliation and branching.
Recorded descriptors were subjected to principal component analysis and average linage hierarchical method of cluster analysis to determine the common pattern of variation among the accessions using SAS version 9.2 (2008) [8].Genetic divergences between clusters were calculated using Mahalanobis [9] and clustering of accessions was done according to Tocher’s method as described by Rao [10].
Result and Discussion
The principal component analysis revealed that the majority of the total variation was contributed by component one and two (Figure 1). Principal component one and two contributed 35% and 19% of the total variation (Table 1) respectively. Maximum genetic variance was contributed by length of basal leaves (0.51) and foliation of the plant (0.49) to principal component 1 while number of basal leaf (0.62) and blade shape of the upper stem leaves (0.54) contributed to principal component two.
| Characters | PC1 | PC2 |
|---|---|---|
| Eigenvalues | 2.79 | 1.5 |
| Proportion of variance | 0.400.21 | |
| Cumulative variance | 0.400.61 | |
| Factor loadings | ||
| Number of Basal Leaf | 0.22 | 0.6 |
| Length of Basal Leaves | 0.51 | 0.22 |
| Length of the Longest Basal Leaf | 0.32 | 0.28 |
| Habitus of the Basal Leaves | -0.46 | |
| Blade Shape of the Longest Basal Leaves | -0.510.25 | |
| Blade Shape of the Upper Stem Leaves | -0.200.54 | |
| Foliation of the plant | 0.49-0.32 | |
| Branching of the plant | 0.00 0.00 | |
Table 1: Principal component analysis of 25 coriander accessions.
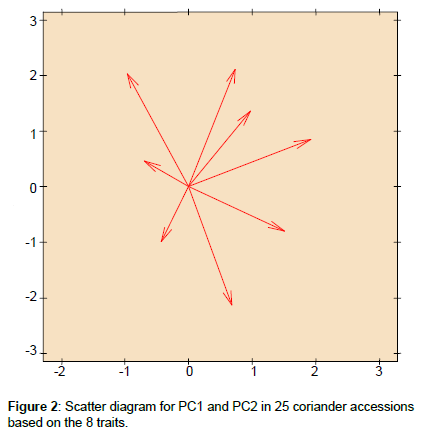
Figure 2 displays a biplot in the dimension of the first and second principal components of the 25 accessions of coriander. Accession 16 and 8 had positive values for PC1 and PC2 and were of considerable breeding interest because of their good combination for the studied traits. Hybridization between the accessions with high positive values for PC1 and close to 0 values for PC2 as accession 24 and 25 and accessions with high positive values for PC2 and near to 0 values for PC1 as accession 22 is expected to give promising and desirable segregates in subsequent generations.
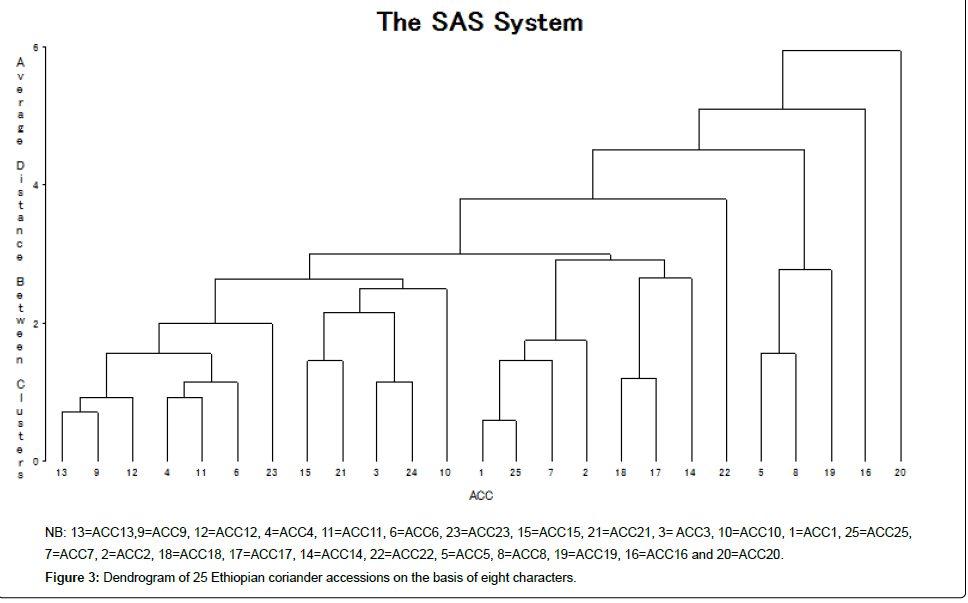
The intra- and inter-cluster distance (D2) values are presented in Table 2. The intra-cluster distance was lower than the inter-cluster distances implying that the accessions included within a cluster had less diversity among themselves. The highest intra-cluster distance (5.67) was observed in cluster I followed by cluster II (1.33). The intracluster distance of cluster III, IV and V was zero for they contained only one accession. The intra-cluster distance in this study is similar to intra-cluster distance (2.81 to 5.59) recorded by Mengesha et al. [3], but relatively lower than the values between 13.8 and 28.25 reported by Singh et al. [1]. The magnitudes of inter-cluster distance (D2) were generally high and were indicator for the presence of substantial genetic diversity in Ethiopian coriander accessions. The highest inter-cluster distance (47.42) was observed between cluster IV and V followed by cluster II and IV (47.33) and cluster I and IV (41.47) suggesting more variability in genetic makeup of the accessions included in these clusters. The accessions belonging to the clusters separated by high statistical distance could be used in hybridization programme for obtaining a wide spectrum of variation among the segregates. In the present study, the range of inter-cluster D2 values from 13.22 (cluster I and III) to 47.42 (cluster IV and V) was obtained. It is comparable to range of inter-cluster distance (13.8 to 91.3) reported by Singh et al. [1]. The 25 accessions were grouped into five clusters based on the 8 characters (Table 3 and Figure 3). Cluster I was the largest having 19 accessions. Cluster II and III had three and two accessions respectively. Cluster IV and V had one accession each.
| I | II | III | IV | V | |
| :I | 5.67 | 15.22 | 13.22 | 41.47 | 18.04 |
| II | 1.33 | 13.72 | 47.33 | 21.74 | |
| III | 0.00 | 39.60 | 28.18 | ||
| IV | 0.00 | 47.42 | |||
| V | 0.00 |
Table 2: Intra-cluster (bolded diagonal) and inter-cluster (off diagonal) distance (D2) values among 25 Ethiopian coriander.
| Cluster | Number of accessions | Accessions |
|---|---|---|
| I | 19 | ACC13, ACC9, ACC12, ACC4,ACC11, ACC6, ACC23, ACC15,ACC21, ACC3, ACC10,ACC1, ACC25,ACC7,ACC2, ACC18, ACC17 and ACC1. |
| II | 3 (5,8,19) | ACC5, ACC8 and ACC19 |
| III | 2 (22)w | ACC22 |
| IV | 1 (16) | ACC16 |
| V | 1 (20) | ACC20 |
Table 3: Clustering pattern of 25 Ethiopian coriander accessions on the basis of 8 traits.
The cluster means of the investigated traits are presented in Table 4. Comparison of means of various traits in different clusters revealed that cluster IV produced the highest mean value (55) for number of basal leaf, length of basal leaf (45.8cm) and length of the longest basal leaf (52 cm) than the other clusters. The same branching habit per plant was observed in all clusters. Cluster I (19 accessions), cluster III (single accessions) and IV (single accession) showed similar blade shape of the longest basal leaf and foliation per plant. That means, most of the accessions (22) evaluated had very many leaves. On the other hand, cluster II, with 3 accessions, had produced middle number of leaves while cluster V, with single accession, had very few leaves. It can be concluded from the finding of the foliation that the clusters with very many leaves can be produced for fresh herb purposes on top of their production for fruit.
| Cluster | NBL | LBL | LLBL | HBL | BSLBL | BSUSL | FOLN | BR |
|---|---|---|---|---|---|---|---|---|
| I | 15.63 | 36.55 | 42.95 | 2 | 4 | 4 | 9 | 1 |
| II | 16.67 | 27.75 | 31.33 | 2 | 5 | 5 | 5 | 1 |
| III | 22 | 39.6 | 32 | 1 | 4 | 6 | 9 | 1 |
| IV | 55 | 45.8 | 52 | 1 | 4 | 5 | 9 | 1 |
| V | 14 | 23.6 | 52 | 4 | 5 | 6 | 1 | 1 |
Note: NBL=number of basal leaf per pant, LBL=length of basal leaf, LLBL=length of the longest basal leaf, HBL=habitus of the basal leaf, BSLBL=blade shape of the longest basal leaf, BSUSL=blade shape of the upper stem leaf, FOLN=foliation and BR=branching of the plant.
Table 4: Cluster mean values of 8 traits of 25 Ethiopian coriander accessions.
On the other and Cluster II and IV and cluster III and V showed similar blade shape of the upper stem leaf. Cluster IV and V, each with single accession, gave the maximum length (52cm) of the longest basal leaf. Cluster II, with three accessions, gave the lowest length (31.33cm) of the longest basal leaf while cluster V, with single accession, produced the lowest length of the basal leaf. Cluster III and IV had very flat (1) while cluster I and II showed flat (2) habitus of the basal leaf. On the other hand, cluster V that had only one accession showed a different leaf habitus that is raised with an arcus of about 45°. Cluster V produced the lowest number of basal leaf (14) per plant.
References
- Singh SP, Katiyar RS, Rai SK, Tripayhi SM, Srivastava JP (2005) Genetic divergence and its implication in breeding of desired plant type in coriander (Coriandrum sativum L.). Genetika 37: 155-163.
- Diederichsen A (1996) Coriander (Coriandrum sativum L.): Promoting the conservation and use of underutilized and neglected crops. Institute of Plant Genetics and Crop Plant Research, Gatersleben/ International Plant Genetic Resources Institute, Rome.
- Mengesha B, Alemaw G, Tesfaye B (2011) Genetic divergence in Ethiopian coriander accessions and its implication in breeding of desired plant types. Afr Crop Sci J 19: 39-47.
- Sirohi SPS, Dar AN (2009) Genetic divergence in soybean (Glycine max L. Mrrill). SKUAST Journal of Research 11: 200-203.
- Marker S, Krupakar A (2009) Genetic divergence in exotic maize germplasm (Zeamays L.). Journal of Agricultural and Biological Science 4: 44-47.
- Palomino EC, Mori ES, Zimback L, Tambarussi EV, Moracs CB (2005) Genetic diversity of common bean genotypes of Carioca commercial group using RADP markers. Crop Breeding and Applied Biotechnology 5: 80-85.
- Dyulgerov N, Dyulgerova B (2013) Variation of yield components in coriander (Coriandrum sativum L.). Agricultural Science and Technology 5: 160-163.
- SARC (1998) A decade of research experience. Geremew E Tilahun G Aliy H (eds.). Bulletin No. 4. Sinana agricultural research center, agricultural research coordination service, Oromia agricultural development bureau.
- Mahalanobis PC (1936). On the generalized distance in statistics. Proceedings of National Institute of Science, India 2: 49-55.
- Rao CR (1952) Advance statistical methods in biometrical research. John Wiley and Sons, New York, 390.
Citation: Fufa M (2013) Genetic Divergence in Ethiopian Coriander (Coriandrum sativum l.) Accessions. Adv Crop Sci Tech 1:116. DOI: 10.4172/2329-8863.1000116
Copyright: © 2013 Fufa M. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 16988
- [From(publication date): 11-2013 - Apr 03, 2025]
- Breakdown by view type
- HTML page views: 12200
- PDF downloads: 4788