Genetic Divergence and Principal Component Analysis of Soybean Glycine max L Merrill Genotypes in Northwestern Ethiopia
Received: 01-Aug-2023 / Manuscript No. acst-23-107881 / Editor assigned: 03-Aug-2023 / PreQC No. acst-23-107881(PQ) / Reviewed: 21-Aug-2023 / QC No. acst-23-107881 / Revised: 26-Aug-2023 / Manuscript No. acst-23-107881(R) / Published Date: 31-Aug-2023 DOI: 10.4172/2329-8863.1000609
Abstract
The volume of Soybean production and productivity in Ethiopia is low as compared to the world average potential due to less diversified soybean genotypes. Despite the increment of soybean production and productivity in national level, it remains low as compared to the world average potential. The reason behind is due to a lack of diversified soybean genotype access and released varieties genetic potential reductions. Hence, genotypes which have not been characterized clustered and tested for their variability subjected for this study. About Eighty-one (81) genotypes were tested in a 9*9 simple lattice design for their variability and relation of among traits using yield and yield related traits, qualitative, and quality traits at Pawe Agricultural Research Center main station and Dibate substation in 2018-2019 cropping season. The analysis of variance revealed that all traits except number of nodules per plant, number of pods plant-1 and number of seeds pod-1 showed highly significant (p<0.01) differences at both tested locations. Sixty three and sixty five percent of variations, from the total, were observed from the 1st 4 PCAs for Pawe and Dibate, respectively. Cluster analysis showed about four different clusters and the maximum inter cluster distance was observed between cluster I and cluster IV (D2=875.31) at Pawe and between cluster II and cluster IV (D2=1227.68) at Dibate. Though the experiment conducted at two locations genotypes variableness would not be realized in a single season since most of the traits are profoundly polygenic. In this regard, additional testing in different seasons and more locations is required.
Keywords
Cluster distance; Genetic divergence; Genetic variability; Genotypes; Principal component; Soybean
Introductio n
Soybean (Glycine max (L.) Merrill) is a self-pollinated and leguminous crop with a chromosome number of 2n = 4x = 40 [1] [2]. Soybean is the most widely grown leguminous crop in the world and is an important source of protein and oil [3][4] and also rich in unsaturated fatty acids, minerals (such as Calcium and Potassium) and vitamins which meet the nutritional needs of humans and other animals [5].
Successful crop improvement programs are critically subjected to the presence of high genetic diversity [6]. In addition to this, genetic diversity enhances the possibility of any species’ existence and being adaptable to fluctuating environmental situations [7, 8]. Hence, loss of genetic diversity puts plants at risk of disease and adverse climate change [9]. Therefore, accurate knowledge of the nature and level of genetic diversity present in soybean germplasms can help in selecting parents to develop the best varieties.
Applications such as the study of genetic divergence between genotypes which permits the identification and selection of the most promising genotypes for cultivation and improvement, and evaluating the relative importance of characters in the total variation available among genotypes are estimated through principal component analysis (PCA) [10].
Despite the increment of soybean production and productivity from 15824.4 tons with a productivity of 1.4 tons ha-1 in 2010 [11] to 86467.9 tons with a productivity of 2.27 tons ha-1 in 2017 [12] in national level, it remains low compared to the world average productivity potential of 2.7 tons ha-1 [12]. The reason behind is due to a lack of diversified soybean genotype access and released varieties genetic potential reductions [13]. On the other hand, increasing of population growth, agro-processing factories and urbanization have resulted in soybean raw materials and products [14]. Hence, genotypes have never yet characterized systematically need to be tested for its variableness since it is influenced by environmental factors [15].
Materials and Methods
Description of experimental sites
The experiment was conducted at Pawe Agricultural Research Center main station and Dibate sub-station in 2018/19. Pawe Agricultural Research Center is located at (11018`49.6``N and 036024`29.1``E) in Metekel Zone. The altitude of the area ranges from 1150 meters above sea level (m.a.s.l). The site receives 1586 mm rainfall annually. The mean annual maximum and minimum temperatures are 32.60c and 16.50c, respectively. The soil type of the site is characterized by well drained clay soil with pH value 4.3-5.5. Dibate substation is located at (10o30′ 0′ N, 36o 10′ 0′ E) with an altitude of 1572 m.a.s.l. The mean annual maximum and minimum temperatures are 29oc and 15oc, respectively and it receives 1650 to 1700 mm rainfall annually. The soil type of the substation is characterized by nitosol or loam type.
Plant materials and experimental design
Eighty-one introduced soybean genotypes from different sources (IITA, USA and Brazil) were used for the experiment (Table 1). The experiment was laid out in 9*9 simple lattice designs with plot size of 7.2 meter square (2.4m*3m). Each plot consisted of four rows with 60 cm inter row and 5 cm intra row spacing. The spacing between plots, blocks and replications were 0.8 m, 1 m and 2 m, respectively. The total net harvestable experimental area for each location was 583.2 m2. The amounts of seed and DAP fertilizer rate per plot were 54g and 72g, respectively (Table 1).
| 1 | Genotype | Source | Year of | S | Genotype | Source | Year of |
|---|---|---|---|---|---|---|---|
| Designation | Introduction | no. | Designation | introduction | |||
| 1 | Tgx-1448-2e | IITA | 2016 | 45 | Tgx-1989-42f | IITA | 2014 |
| 2 | Tgx-2010-11f | IITA | 2015 | 46 | Tgx-1989-45f | IITA | 2014 |
| 3 | Tgx-1989-19f | IITA | 2015 | 47 | Tgx-1989-75f | IITA | 2014 |
| 4 | Tgx-2006-3f | IITA | 2015 | 48 | Tgx-1990-106fn | IITA | 2014 |
| 5 | Tgx-2008-4f | IITA | 2016 | 49 | Tgx-1990-107fn | IITA | 2014 |
| 6 | Tgx-2010-12f | IITA | 2016 | 50 | Tgx-1990-110fn | IITA | 2014 |
| 7 | Tgx-2004-10f | IITA | 2016 | 51 | Tgx-1990-111fn | IITA | 2014 |
| 8 | Tgx-1485-1d | IITA | 2016 | 52 | Tgx-1990-114fn | IITA | 2014 |
| 9 | Tgx-2007-8f | IITA | 2016 | 53 | Tgx-1990-87f | IITA | 2014 |
| 10 | Tgx-2008-2f | IITA | 2016 | 54 | Tgx-1990-8f | IITA | 2014 |
| 11 | Tgx-2004-13f | IITA | 2016 | 55 | Tgx-1990-95f | IITA | 2014 |
| 12 | Tgx-2007-11f | IITA | 2016 | 56 | Tgx-1993-4fn | IITA | 2014 |
| 13 | Tgx-2010-15f | IITA | 2016 | 57 | Tgx-1989-48fn | IITA | 2014 |
| 14 | Tgx-2011-3f | IITA | 2016 | 58 | Tgx-1989-68f | IITA | 2014 |
| 15 | Tgx-2010-3f | IITA | 2016 | 59 | Tgx-1990-78f | IITA | 2015 |
| 16 | Tgx-2004-3f | IITA | 2016 | 60 | Tgx-1990-57f | IITA | 2014 |
| 17 | Tgx-2011-7f | IITA | 2016 | 61 | Tgx-1835-10f | IITA | 2014 |
| 18 | Tgx-1987-10f | IITA | 2016 | 62 | Tgx-1995-5f | IITA | 2014 |
| 19 | Tgx-1987-42f | IITA | 2016 | 63 | Tgx-1987-20f | Malawi | 2009 |
| 20 | Tgx-1987-45f | IITA | 2013 | 64 | Tgx-1987-23f | Malawi | 2009 |
| 21 | Tgx-1990-101f | IITA | 2013 | 65 | Tgx-1987-64f | Malawi | 2009 |
| 22 | Tgx-1990-47f | IITA | 2013 | 66 | Tgx-1987-65f | Malawi | 2009 |
| 23 | Tgx-1990-70f | IITA | 2013 | 67 | Tgx-1987-6f | Malawi | 2009 |
| 24 | Tgx-1990-73f | IITA | 2013 | 68 | Tgx-1987-37f | Malawi | 2009 |
| 25 | pr142-15-SG | USA | 2016 | 69 | Tgx-1987-35f | Malawi | 2009 |
| 26 | H3-15-SG | USA | 2016 | 70 | Tgx-1987-38f | Malawi | 2009 |
| 27 | H3-15-SE-1 | USA | 2016 | 71 | Tgx-1987-15f | Malawi | 2009 |
| 28 | CLK-15-sb-1 | USA | 2016 | 72 | Tgx-1987-19f | Malawi | 2009 |
| 29 | ALM-15-SB | USA | 2016 | 73 | Tgx-1986-3f | Malawi | 2009 |
| 30 | CRFD-15-SC | USA | 2016 | 74 | Tgx-1987-40f | Malawi | 2009 |
| 31 | PR142-1-SE | USA | 2016 | 75 | Tgx-1740-2f | Malawi | 2009 |
| 32 | G99-15-SE-2 | USA | 2016 | 76 | pb12-1 | Brazil | 2012 |
| 33 | CLK-15-SA-1 | USA | 2016 | 77 | pb12-6 | Brazil | 2012 |
| 34 | CRFD-15-SB | USA | 2016 | 78 | pb12-7 | Brazil | 2012 |
| 35 | CLK-15-SA-1 | USA | 2016 | 79 | pb12-8 | Brazil | 2012 |
| 36 | H3-15-SB-2 | USA | 2016 | 80 | pb12-4 | Brazil | 2012 |
| 37 | G99-15-SA | USA | 2016 | 81 | pun11-4 | Brazil | 2012 |
| 38 | SCS-1 | USA | 2016 | ||||
| 39 | Tgx-1990-78f | IITA | 2013 | ||||
| 40 | Tgx-1990-80f | IITA | 2013 | ||||
| 41 | Tgx-1990-95f | IITA | 2013 | ||||
| 42 | Tgx-1991-10f | IITA | 2013 | ||||
| 43 | Tgx-1904-6f | IITA | 2013 | ||||
| 44 | Tgx-1989-11f | IITA | 2014 |
Table 1: 81 soybean genotypes considered in the experiment.
Data collected
Data on days to flowering, days to maturity, protein and oil contents (in %) and grain yield (kgplot‑1) were recorded on plot bases whereas, number of nodule, number of branches, plant height and number of pods plant-1 were recorded from ten randomly selected plants according to.
Protein and oil quality data were determined by using 150 grams of dried seed samples per genotype and grinded in the quality research laboratory. A small cup with internal diameter of 35 millimeter and depth of 8 millimeter was used to take two to three soybean seed powder. NIRS (Near Infrared spectroscopy) FOSS 6500 model was used for Scanning the oil and protein contents. Global calibration for proximate composition (list of parameters) was predicted as of.
Data analyses
Analysis of variance (ANOVA) was done using proc GLM for the traits analyzed based on RCBD and proc lattice procedures of SAS version 9.3 for the traits analyzed based on lattice.
Cluster analysis and genetic divergence
Cluster analysis was performed using the SAS proc cluster procedure. D square statistics (D2) developed by was used to cluster
genotypes into different groups. The average intra and inter cluster
D2 values were computed by the formulae 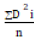 where, ΣD2i stands
the summation of distances between all possible groupings (n) of the
genotypes involved in the group. Genetic difference was estimated by
the generalized Mahalanobis’s statistics
where, ΣD2i stands
the summation of distances between all possible groupings (n) of the
genotypes involved in the group. Genetic difference was estimated by
the generalized Mahalanobis’s statistics 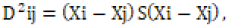 where, D2ij = the distance between two groups i and j. Xi and Xj = the
two vectors mean ith and jth genotypes, respectively; S = is the inverse
of the pooled divergence matrix. The D2 values obtained for sets of
groups were measured as per the computed values of chi‐ square (χ2)
and were tested for significance at (1% and 5%) probability levels
contrary to the tabulated value of χ2 for ‘P’ degree of freedom, where
P stands for the number of parameters considered. Cubic clustering
criteria (CCC), Pseudo F statistic and pseudo t2 statistic generated by
SAS were examined to decide the number of optimum clusters.
where, D2ij = the distance between two groups i and j. Xi and Xj = the
two vectors mean ith and jth genotypes, respectively; S = is the inverse
of the pooled divergence matrix. The D2 values obtained for sets of
groups were measured as per the computed values of chi‐ square (χ2)
and were tested for significance at (1% and 5%) probability levels
contrary to the tabulated value of χ2 for ‘P’ degree of freedom, where
P stands for the number of parameters considered. Cubic clustering
criteria (CCC), Pseudo F statistic and pseudo t2 statistic generated by
SAS were examined to decide the number of optimum clusters.
Principal component analysis
Principal components were estimated based on original data using the SAS PRINCOMP procedure based on formulae given by [23]. The first and second PCAs’ values (Y1) and (Y2) is given by the linear combination of the variables X1, X2…Xp. Y1=a11X1+a12+…a1pXp and Y2=a21X1+a22X2+…a2pXp, respectively. Principal components with Eigen values > 1 considered as a significant in the result.
Results and Discussion
Genetic divergence analysis
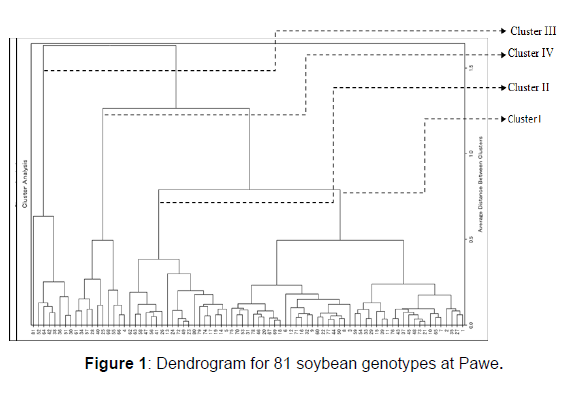
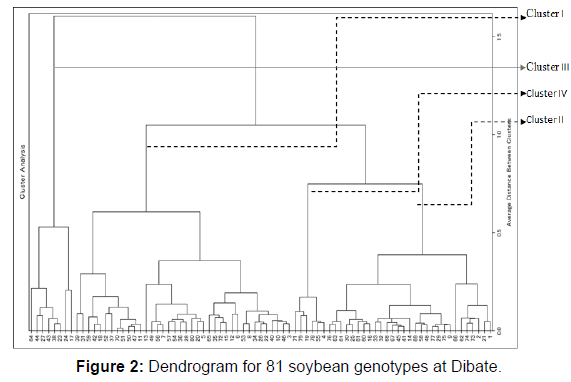
The distribution of genotypes by cluster from the largest to the lowest is cluster I, II, IV and III with 44 (54.3%), 19 genotypes (23.5%), 10 (12.3%) and 8(9.9%) genotypes at Pawe, respectively (Table 2). Whereas at Dibate, cluster I, II, III and IV with 38 (46.9%), 29 (35.8%), III 8 (9.9%) and IV 6 (7.4%) genotypes, respectively (Table 3).
| Cluster | Number of genotypes | Genotypes included |
|---|---|---|
| I | 44 | Tgx-1448-2e, Tgx-2010-11f, Tgx-1989-19f, Tgx-2010-12f, Tgx-2004-10f, Tgx-1485-1d, Tgx-2007-8f,Tgx-2008-2f, Tgx-2004-13f, Tgx-2007-11f, Tgx-2010-3f, Tgx-2004-3f, Tgx-1987-10f, Tgx-1987-45f, Tgx-1990-101f, Tgx-1990-47f, H3-15-SE-1, ALM-15-SB, PR142-1-SE, G99-15-SE-2, CLK-15-SA-1, G99-15-SA, Tgx-1990-78f, Tgx-1904-6f, Tgx-1989-11f, Tgx-1989-42f, Tgx-1989-45f, Tgx-1990-106fn, Tgx-1990-110fn, Tgx-1990-87f, Tgx-1990-8f, Tgx-1990-78f, Tgx-1990-57f, Tgx-1987-6f, Tgx-1987-64f, Tgx-1987-35f, Tgx-1987-38f, Tgx-1987-15f, Tgx-1986-3f, Tgx-1740-2f, pb12-1, pb12-6, pb12-7. |
| II | 19 | Tgx-2008-4f, Tgx-2010-15f, Tgx-2011-3f, Tgx-2011-7f, Tgx-1987-42f, Tgx-1990-70f, Tgx-1990-73f, H3-15-SG, Tgx-1989-75f, Tgx-1990-107fn, Tgx-1993-4fn, Tgx-1989-68f, Tgx-1995-5f, Tgx-1987-20f,Tgx-1987-19f, Tgx-1987-40f, pb12-8, pb12-4. |
| III IV | 8 10 | CRFD-15-SC, H3-15-SB-2, SCS-1, Tgx-1991-10f, Tgx-1990-111fn, Tgx-1990-114fn, Tgx-1987-23f, pun11-4. Tgx-2006-3f, pr142-15-SG, CLK-15-sb-1, CRFD-15-SB, Tgx-1990-80f, Tgx-1990-95f, Tgx-1989-48fn, Tgx-1835-10f, Tgx-1987-65f, Tgx-1987-37f. |
Table 2: The distribution of 81 genotypes based on D2 analysis at Pawe.
| Cluster | Number of genotypes | Genotypes included |
|---|---|---|
| I | 38 | Tgx-1989-19f , Tgx-2010-12f, Tgx-2008-4f, tgx-2004-10f, Tgx-1485-1d, Tgx-2008-2f, Tgx-2004-13f, Tgx-2007-11f, Tgx-2010-15f, Tgx-2010-3f, Tgx-1987-10f, Tgx-1987-45f, Tgx-1990-47f, H3-15-SG, CLK-15-sb-1, PR142-1-SE, CRFD-15-SB, CLK-15-SA-1, H3-15-SB-2, G99-15-SA, Tgx-1990-78f, Tgx-1990-80f, Tgx-1991-10f, Tgx-1989-45f, Tgx-1989-75f, Tgx-1990-107fn, Tgx-1990-110fn, Tgx-1990-111fn, Tgx-1990-114fn, Tgx-1990-87f, Tgx-1990-8f, Tgx-1993-4fn, Tgx-1989-48fn, Tgx-1990-78f, Tgx-1987-64f, Tgx-1987-38f, Tgx-1987-19f, pb12-4. |
| II | 29 | Tgx-1448-2e, Tgx-2010-11f, Tgx-2007-8f, Tgx-2011-3f, Tgx-2004-3f, Tgx-1990-101f, Tgx-1990-73f, ALM-15-SB, CRFD-15-SC, G99-15-SE-2, CLK-15-SA-1, Tgx-1990-95f, Tgx-1989-42f, Tgx-1990-106fn, Tgx-1989-68f, Tgx-1990-57f, Tgx-1835-10f, Tgx-1995-5f, Tgx-1987-20f, Tgx-1987-65f, Tgx-1987-6f, Tgx-1987-37f, Tgx-1987-35f, Tgx-1986-3f, Tgx-1987-40f, Tgx-1740-2f, pb12-1, pb12-6, pun11-4. |
| III | 8 | Tgx-2011-7f, Tgx-1990-70f, pr142-15-SG, H3-15-SE-1, SCS-1, Tgx-1904-6f, Tgx-1989-11f, Tgx-1987-23f. |
| IV | 6 | Tgx-2006-3f, Tgx-1987-42f, Tgx-1990-95f, Tgx-1987-15f, pb12-7, pb12-8. |
Table 3: The distribution of 81 genotypes based on D2 analysis at Dibate.
Cluster distance of soybean genotypes
Cluster I and IV (D2=875.3), Cluster I and III (D2=580.05) and cluster II and IV (D2=529.9) showed the greatest inter cluster distance at Pawe (Table 4). Whereas between clusters II and IV (1227.7), clusters I and II (853.8) and cluster III and cluster IV (779.5) at Dibate (Table 5). The shortest squared distance was found between clusters I and IV (373.9) following by clusters I and III (405.7) and between cluster II cluster III (448.2). As a result, clusters with the largest distance indicate the variableness between genotypes that are included between those clusters. The presence of the genetic distance between clusters maximizes the chance of bread wheat varieties improvement through wide crosses and high expression of heterosis had also reported by. Besides, crossing of genotypes from distant inter clusters may also produce higher range of differences in successive segregate populations.
| Cluster | I | II | III | IV |
|---|---|---|---|---|
| I | 50.9 | 345.5** | 580.05** | 875.31** |
| II | 76.5 | 234.69** | 529.96** | |
| III | 71.7 | 295.29** | ||
| IV | 81 |
Table 4: Intra (bold diagonal) and inter Mahalanobis distance among genotypes at Pawe.
| Cluster | I | II | III | IV |
|---|---|---|---|---|
| I | 102.8 | 853.8** | 405.7** | 373.9** |
| II | 131.7 | 448.2** | 1227.7** | |
| III | 135 | 779.5** | ||
| IV | 111 |
** = significant at 1% levels.
Table 5: Intra (bold diagonal) and inter Mahalanobis distance among genotypes at Dibate.
Thus, crossing between genotypes involving between clusters I and IV at Pawe and between clusters II and IV at Dibate is suggested to reveal better recombinants and could result in segregates with higher seed yield.
Cluster mean analysis
Genotypes with the second highest mean value for protein and the lowest mean value for oil content are indicated in cluster I, whereas the lowest mean values of number of nodule, plant height, number of branches, number of pods, second highest a hundred seed weigh and lowest grain yield genotypes are included in cluster II at Pawe (Table 6). Similarly, cluster characterized by lowest mean values of number of branches, number of pods and grain yield in soybean genotypes has been reported by. Genotypes with the highest means values for days to flowering, days to maturity, number of nodules, plant height, number of branches, second largest mean values for number of pods and grain yield in cluster III and with the lowest mean values for days to flowering, the lowest days to maturity and protein content and highest mean values for number of branch, number of pods, number of seeds, a hundred seed weight and grain yield cluster IV. Such a reverse relationships of days to maturity with yield and yield related traits would be observed in response of disease and moisture stress occurrences. These unfavorable conditions, in fact, were happened during this field experiment due to that planting time was being late. Frogeye leaf spot was also occurred at pod filling stage of the crop especially at Pawe location. These might be caused that, consequently, most of genotypes could not attain their seed as expected. The same finding has been reported by different workers that genotypes with the lowest mean values of days to flowering, days to maturity and with the highest mean value of grain yield in soybean with in a cluster.
At Dibate (Table 7), genotypes are characterized by the second least average values for number of branches, number of pods, number of nodules and grain yield in cluster I; by the least average values for days to flowering, number of nodule, days to maturity, number pods and seeds, a hundred seed weight and yield in cluster II; by the maximum average values for days to maturity , the highest plant height, second largest mean value for number of pods, the highest protein content and second largest yield in cluster III and by the maximum average values for days to flowering, number of nodule, days to maturity, branch number, pod number, seed number, a hundred seed weight and grain yield in cluster IV. Similarly, cluster characterized by highest mean values of days to flowering, days to maturity & grain yield of soybean has been reported by. Genotypes in the second cluster will be used for crossing with genotypes in cluster IV to develop late maturing soybean genotypes with high number of pods, high hundred seed weight and grain yield (Tables 2&3) (Figures 1&2) (Tables 4-7).
| Cluster | ||||
|---|---|---|---|---|
| Traits | I | II | III | IV |
| DF | 69 | 68 | 70 | 70.5 |
| NN | 12.6 | 12.2 | 14.1 | 16.3 |
| DM | 124 | 123 | 126 | 125.2 |
| PH | 62.6 | 59.9 | 63.6 | 61.4 |
| BrP | 4 | 4.1 | 4 | 4.4 |
| PdP | 30.9 | 28.7 | 32.3 | 36.4 |
| SdP | 1.78 | 1.7 | 1.8 | 1.8 |
| HSW | 12.2 | 11 | 12.1 | 13 |
| Oil | 21 | 20.6 | 20.7 | 20.5 |
| Protein | 36 | 36 | 36.6 | 36.3 |
| Yield | 1484.6 | 1110.4 | 1842.9 | 2324.2 |
Table 7: Cluster mean for 11 traits in soybean tested at Dibate.
DF=days to 50% flowering, NN=number of nodule per plant, DM=days to 95% maturity, PH= plant height, BrP= number of branches per plant, PdP=number of pods per plant, SdP=number of seeds per pod, HSW=hundred seed weight.
Principal components analysis
The principal components which had the highest contribution for the total variability of the eighty-one genotypes are given in Table 6. Variations explained by the first four PCAs from total variations were 63% and 65% at Pawe and Dibate, respectively. In the first principal components, traits with positive and high value were days to flowering (0.48), days to maturity (0.44), plant height (0.33) and protein content (0.30) at Pawe and days to maturity (0.46), plant height (0.42), protein content (0.38) and days to flowering (0.36) at Dibate.
| Clusters | ||||
|---|---|---|---|---|
| Traits | I | II | III | IV |
| DF | 58.1 | 56.9 | 59.4 | 53.2 |
| DM | 114.1 | 111.4 | 114.5 | 113.2 |
| NN | 17.4 | 16.5 | 18.2 | 17.4 |
| PH | 82 | 76.2 | 82.1 | 76.7 |
| BrP | 4.2 | 3.9 | 4.5 | 4.5 |
| PdP | 53.9 | 48.7 | 60.1 | 64.2 |
| SdP | 1.92 | 1.91 | 1.87 | 2.1 |
| HSW | 12.7 | 13.1 | 12.7 | 13.9 |
| Protein | 35.7 | 35.9 | 35.9 | 34.2 |
| Oil | 21 | 21.4 | 21.1 | 22 |
| Yield | 2094.4 | 1812.1 | 2361.3 | 2631.4 |
Table 6: Cluster mean for eleven traits in soybean traits at Pawe.
The major contributing traits in the second principal component with high and positive component loading were number of branch (0.51), number of pods (0.5) and plant height (0.29) at Pawe and number of branch (0.6), pod number (0.46) and oil content (0.31) at Dibate. Traits with high and positive component loading were yield (0.59) and number of nodule (0.47) from the third PCA at Pawe and number of seeds (0.61), nodule number (0.47) and a hundred seed weight (0.38) at Dibate. The same finding reported that numbers of pods per plant, total seeds per pod & grain yield were the major contributing traits in PCA of soybean genotypes. Traits loaded with high positive or negative values accounted more to the variability and they considered that are the most differentiated the cluster [30] (Table 8). PCs= Principal components, DF=Days to 50% flowering, NN=Number of nodules per plant, DM= Days to 95% maturity, PH=Plant height, BrP= number of branches per plant, PdP=Number of pods per plant, SdP=number of seeds per pod and HSW= Hundred seed weight.
Conclusions
The soybean genotypes grouped into four distinct clusters at both locations. The first four PCAs were found to be significant (with Eigen values greater than one) and accounted for about 63% and 65% from the total variation at Pawe and Dibate, respectively.
Genotypes from distant inter cluster (cluster I and cluster IV) at Pawe and cluster II and IV (1227.7) at Dibate and genotypes with major variability contributor traits having high value such as days to flowering, days to maturity, plant height and protein content at both locations from the first principal components will be used as a parental material for crossing.
However, one season experiment would not realize genotypes’ variability in response of environment, because quantitative traits are polygenic and profoundly influenced by the environment. Thus, further experiment on these genotypes in changed seasons is required.
Acknowledgments
I forward my deepest thanks to Ethiopian Institute of Agricultural Research which gave me the study grant and leave. I express my exceptional appreciations to Pawe Agricultural Research Center, intended for its easing of my thesis research, by utilization of properties, organizational support and delivery of testing plots and other needed supplies.
Conflict of Interest
Writers ensure that not any opposing profits occur. The materials and inputs used for this research are commonly and predominantly from the employer institution in I am working for. There is absolutely not any clash of importance between the authors and of the institution. Also, the research was funded only by the institution.
References
- Hymowitz T (2004) Speciation and cytogenetic Soybeans Improvement, Production, and Uses. American Society of Agronomy Crop Science Society of America Soil Science Society of America Madison Wis 97–136.
- Smith KJ, Huyser W (1987) World Distribution and Significance of Soybean. American Society of Agronomy Inc. Madison WI 1-22.
- Sharma S, Kaur M, Goyal R, Gill BS (2013) Physical characteristics and nutritional compositions of some new soybean Glycine max L. Merrill genotypes. Journal of food science and Technology 51(8): 551-557.
- Hossain Z, Komatsu S (2014) Potentiality of soybean proteomics in untying the mechanism of Flood and Drought stresses tolerance. Proteomes 2(1): 10-127.
- Ghosh J, Ghosh PD, Choudhury PR (2014) an assessment of genetic relatedness between soybeans cultivars using SSR markers. American journal of plant Sciences 5(1): 3089-3096.
- Salgotra RK, Gupta BB, Bhat JA, Sharma S (2015) Genetic diversity and population structure of basmati rice Oryza sativa L germplasm collected from north western Himalayas using trait linked SSR markers. 10(7): 1-19.
- Otálora MA, Belinchón R, Prieto M, Aragón G, Izquierdo P, et al (2015) The Threatened Epiphytic lichen lobaria Pulmonaria in the Iberian Penisula. Genetic Diversity and Structure across a latitudinal gradient. Fungal 119(9): 802–811.
- Chen YY, Wang WC, Fan XR, Sun JY, Li W, et al (2019). Genetic discontinuities and abundant historical gene flow in wild lotus Nelumbo Nucifera Population from the Yangtze River. Aquat Bot 158: 103130.
- Punia SS, Baldev R, Koli NR, Ranwah BR, Rokadia P, et al (2011) Genetic architecture of quantitative traits in field pea. Journal of Swollen Food 24(6): 299-303.
- Vianna VF, De Santiago S, Charnai K, Ferraudo AS, Di Mauro AO (2013) The multivariate approach and Influence of Characters in Selecting superior Soybean genotypes. Afr J Agric Res 8(30): 4162–4169.
- Central Statistical Agency of Federal Democratic Republic of Ethiopia. Annual report 2000-2012.
- Central Statistical Authority (CSA) (2018). Agricultural sample survey. Addis Ababa Ethiopia. 1: 14.
- Mesfin HM, Abush T (2018) Progress of Soybean Glycine max Merrill L Breeding and Genetics Research in Ethiopia. Journal of Natural Sciences Research 8:70.
- Afework H, Adam B (2018) Cost and returns of soybean production in Assosa Zone of Benishangul Gumuz Region of Ethiopia. Journal of Development and Agricultural Economics 10(11): 377-383.
- Dudley JW (1993) Molecular Markers in Plant plant improvements manipulation of genes affecting quantitative traits. Crop Sci 33(8): 660–668.
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Citation: Mekonen AA (2023) Mungbean Vigna radiata (L.) Wilczek Varieties Evaluation Trial Based on Some Selected Physiological Growth Parameters at Hawassa Ethiopia. Adv Crop Sci Tech 11: 609. DOI: 10.4172/2329-8863.1000609
Copyright: © 2023 Mekonen AA. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 1504
- [From(publication date): 0-2023 - Apr 04, 2025]
- Breakdown by view type
- HTML page views: 1268
- PDF downloads: 236