Functional Differences in Pertussis Toxins from Bordetella pertussis Clinical Isolates as Determined by in vitro and in vivo Assays
Received: 29-Jun-2015 / Accepted Date: 28-Jul-2015 / Published Date: 30-Jul-2015 DOI: 10.4172/2161-0681.1000243
Abstract
Whooping cough caused by Bordetella pertussis is a serious disease especially for infants and young children. Detoxified pertussis toxin is a key component of vaccines used in campaigns worldwide for the prevention of the disease. A biochemical assay system, which measures pertussis toxin enzymatic and carbohydrate binding activities, has been developed to measure the residual toxin activities in vaccine matrix. We report here that B. pertussis clinical isolates show differences in pertussis toxin (PTx) activities in the in vitro biochemical assays and these in vitro activities were also positively related to the in vivo toxic activities. In addition, interesting information on the genetic and possible post-translational changes of PTx produced by the strains included in this study is tentatively discussed. Of the six strains studied in details, three low- and three high-activity strains, all the DNA sequences of the ptx gene clusters were found to be identical, except that in the high-activity strains, there was a silent mutation with a single nucleotide change at base 681 in ptxC which coded for Cys199 of PTx subunit S3. Mass spectrometric analysis of tryptic peptides from PTx produced by these strains detected two peptides in subunits S1 and S3, which were present in the low-activity strains but not in the high-activity strains. This suggests that PTx might differ in structure between these strains and this is possibly due to post-translational modification.
Keywords: B. pertussis; Strains; Pertussis toxin; Binding; Enzyme; Histamine-sensitisation; Lymphocytosis
320094Abbreviations
aP: Acellular Pertussis; ACV: Acellular Pertussis Vaccines; AEBSF: 4-(2-Aminoethyl) Benzenesulfonyl Fluoride Hydrochloride; AGP: Human Plasma Alpha-1 Acid Glycoprotein; EDQM: European Directorate for the Quality of Medicines and Healthcare; Fet: Fetuin; FHA: Filamentous Haemagglutinin; Fim2 and Fim3: Fimbriae type 2 and type 3; HIST: Histamine-Sensitisation Test; hTf: Human Transferrin; i.p: Intra-Peritoneally; NIBSC: National Institute for Biological Standards and Control; Prn: Pertactin; ptx: Pertussis Toxin Allele; PTx: Pertussis Toxin; ptxA: PTx Subunit S1 Allele; ptxC: PTx Subunit S3 Allele; PTd: Pertussis Toxoid; PBSG: Phosphate Buffered Saline Containing 0.2% (w/v) Gelatine; PFGE: Pulsed Field Gel Electrophoresis; RNaseB; Bovine Pancreas Ribonuclease B; WCV: Whole-Cell Pertussis Vaccines
Introduction
Pertussis (whooping cough) is still a major cause of morbidity and mortality, especially in infants and vaccination against Bordetella pertussis is a key component of immunisation campaigns worldwide for the prevention of the disease. Pertussis toxin (PTx) is a protein toxin which has a wide range of biological activities in vivo including induction of lymphocytosis, histamine-sensitisation, increased insulin production with consequent hypoglycaemia, potentiation of anaphylaxis and lethality in mice and playing a possible role in neuropathology [1-3]. Excessive residual PTx activity in acellular pertussis (aP) combination vaccines that contain diphtheria and tetanus toxoids (DTaP) could also cause intensified sensitisation to components with subsequent severe local reaction to a booster dose in an animal model [4]. Although these biological effects of PTx have been under extensive research, its mechanism(s) of toxicity is still unclear. Two in vivo assay procedures are commonly used for assessing toxicity of PTx: (1) mouse lymphocytosis test which is based on the fact that the severity of pertussis infection in infants was found to accompany with extremely high lymphocyte counts and this characteristic has been attributed to the PTx [5]; (2) mouse histamine sensitisation test (HIST) where PTx, when administered in vivo, enhances vascular permeability and therefore is capable of inducing vasoactive amine-sensitising activities which can result in death due to hypotensive and hypovolemic shock following vasoactive amine (e.g. histamine) challenge of PTx-treated animals. HIST is currently the accepted WHO and European Pharmacopoeia safety test for determination of PTx activity in pertussis vaccines [6,7].
PTx has the A-B5 type structure typical of many other bacterial toxins, having an enzymatically active A-protomer, the monomeric subunit S1, and a host cell binding B-oligomer which is composed of subunits S2 through to S5 [8,9]. Intact B-oligomer is required for the binding of the holotoxin to receptor sites on the cell surface and enables entry of the A-protomer into the cells [9,10]. The translocated PTx A-protomer catalyses ADP-ribosylation of the α-subunit of eukaryote GTP-binding regulatory proteins, which results in the prevention of the hormonal inhibition of adenylate cyclase and subsequently, an increase in the intracellular levels of cAMP and cell death [10]. Recently, a biochemical in vitro assay system has been developed to measure the residual PTx toxicity in pertussis toxoid (PTd) and pertussis components containing vaccines [11]. The enzymatic activity of PTx A-protomer can be assayed by measuring ADP-ribosylation of a fluorescent synthetic peptide substrate using an enzyme-HPLC coupled assay (E-HPLC) [12,13], while the binding activity of B-oligomer can be measured by a carbohydrate binding assay [14]. The relevance of this test system to some of the physio-pathological effects of PT in vivo has already been described [15,16].
Following the introduction of B. pertussis vaccination with whole cell vaccines (WCV) in the 1950s, the pertussis vaccine has made a major contribution to decreasing the incidence of pertussis disease [17]. However, pertussis still remains endemic worldwide and is an important public health problem. Over the past decade, there has been a resurgence of reported pertussis cases in many regions of the world where the acellular pertussis vaccination coverage in young children is high, e.g. Netherlands, Canada, USA, France and UK [18-20]. Waning immunity and pathogen adaptation, involving antigenic divergence between clinical isolates and vaccine strains, were proposed to be the causes for the resurgence [18,19,21,22]. Recently epidemiological data suggest more virulent B. pertussis strains associated with increased notification in the Netherlands, Sweden and Australia have emerged [23-25]. So far, the relative toxic activity of PTx produced by clinically isolated strains has not been reported. In order to further understand the mechanism of virulence in clinical isolates, in the present study we measured their in vitro enzymatic and carbohydrate binding activities, in vivo toxic activities and analysed the genetic and amino acids sequences of PTx from clinical isolates.
Materials and methods
Bacterial strains
A total of 31 clinical isolates (17 ptxP1 and 14 ptxP3) collected in Finland and The Netherlands during the period of 1999-2004 [23,26] were first screened for PTx activity by the in vitro biochemical methods.
For investigation of the relationship of the in vitro bioactivities and the in vivo toxic activities of these strains, due to ethical reasons on animal experimentation, only three strains expressing the lowest and three strains with the highest in vitro PTx activities (refer as low- and high-activity in the text) were selected to perform in vivo toxicity studies (Table 1). The selected six strains had the same genotypes for PTx subunit S1 (ptxA) and pertactin. The PFGE (pulsed field gel electrophoresis) cluster characterisation of the low-activity strains belongs to IVγ and one of the high-activity strains belongs to cluster IVβ [27] while the PFGE type of the other two strains were not known. Strains were grown on charcoal agar supplemented with 10% horse blood and incubated for 2 days at 37°C and sub-cultured once under the same culture condition for 18 h.
| ptxP | ptxA | prn | Serotype | PFGE cluster | |
|---|---|---|---|---|---|
| PRCB12 | 1 | 1 | 2 | 2 | IVγ |
| PRCB20 | 1 | 1 | 2 | 2 | IVγ |
| PRCB309 | 1 | 1 | 2 | 2 | IVγ |
| PRCB529 | 3 | 1 | 2 | 3 | IVβ |
| B1865 | 3 | 1 | 2 | 3 | nk |
| B2903 | 3 | 1 | 2 | 2,3 | nk |
Table 1: Characteristics of the B. pertussis strains included in the study [1].
Preparation of bacterial cell supernatant and purification of PTx
Bacteria collected from charcoal agar plates were suspended into sterile PBS containing 2 mM protease inhibitor (AEBSF: Sigma, A8456). The suspension was sonicated for 2 × 5minutes in a sonic water bath and was then centrifuged for 5 minutes at 5000 rpm to remove cell debris. For each strain tested, 2-3 independent cultures were prepared and their individual supernatants were collected for analysis. Total protein concentration (mg/ml) in supernatant was determined by UV at 275 nm (Perkin Elmer Lambda 800 UV/Vis spectrophotometer) against a bovine serum albumin standard.
For proteomic analysis, bacterial cultures were grown in Stainer and Scholte liquid media for 24 h and supernatant was used for the purification of PTx according to the method described by Sato et al [28] and Pourahmadi, et al [29]. In brief, protein solution (2-4 mg) was loaded onto a hydroxyapatite column and PTx was eluted with 100 mM potassium phosphate (pH 8) buffer. PTx in the elution fractions were identified by PTx specific sandwich ELISA (see below) and those fractions with highest concentrations were pooled. The pooled PTx was concentrated and further purified by affinity absorption onto fetuin bound magnetic beads (Thermo Scientific, Cat # 88826) which were prepared according to the manufacturer’s protocol. Samples and beads were allowed to mix on a shaker (600 rpm) at 4°C overnight and magnetic beads were separated by placing on a magnetic stand for 3-5 min. After the removal of supernatant, beads were washed twice with Tris buffered saline (0.05M Tris/HCl, 0.15M NaCl, pH7.6) and bound PTx was eluted from the beads using elution buffer (4 M MgCl2, 0.05 M Tris/HCl, pH 7.4). The concentration of PTx was determined by a PTx specific sandwich ELISA.
Determination of concentration of PTx
Immune-dot blotting and sandwich ELISA methods were used for determining the PTx concentration. PTx reference, NIBSC 90/518 [30] was included in each assay as reference for calculation purposes. The immune-dot blotting assay was performed similar to the method described by Stott [31]. In brief, samples and the reference were serially diluted and 200 µl/well was transferred to the dot-blot apparatus containing nitrocellulose membrane (Amersham Life Sciences, RPN203G). After the samples had drained through, the membrane was removed, washed and blocked with 5% (w/v) Marvel skimmed milk in PBS-0.05% (v/v) Tween-20 for 1 h at 37°C. After washing, the membrane was treated with anti-PTx sheep serum (NIBSC, 97/572) for 2 h at 37°C. After washing, the membrane was treated with anti-sheep HRP-conjugate (Sigma, A3415) for 2h at 37°C and then treated with 3,3’-diaminobenzidine substrate solution (Sigma, D4418) for 15 min according to the manufacturer’s instruction. The coloured dot density was measured under white light on a bio-imaging system (UVP Lab Products, UK) and estimated by the Lab Works computer software (Version 3.0.02.00). Parallel line analysis was carried out using the CombiStats software (Version 4.0, EDQM) to calculate the concentration of PTx against the reference.
The PTx specific sandwich ELISA was performed according to the method described by Sato et al. [32]. In brief, 96-well plate wells were coated with monoclonal antibody (Mab) to PTx subunit S1 (NIBSC code 99/506) overnight at 37°C. After washing and blocking, samples and the reference were added to the coated plate and serially diluted and incubated at room temperature for 1.5 h. After washing, guinea-pig antiserum to PTx (in-house reagent) was added into the plate and incubated at room temperature for 1.5 h. After washing, 100 μl of anti-guinea pig IgG HRP-conjugate was added to each well and incubated for 1.5 h. OPD peroxidase substrate (Sigma, P9187) solution was then added to each well and incubated for 10-15 min in the dark. The reaction was stopped by the addition of 50 μl of 3 M HCl to each well and the absorbance at 492 nm was measured in an ELISA plate reader (Labsystem Multiscan MS, UK). Parallel line analysis was carried out using the CombiStats software (Version 4.0, EDQM) to calculate the concentration of PTx against the reference.
Enzymatic-HPLC coupled assay and carbohydrate-binding assay
The determination of PTx ADP-ribosylation activity in bacterial cell supernatant was performed using a synthetic fluorescein-tagged peptide substrate, F-Gαi3C20 (Cat #HCAM-2; AnaSpec Inc., San Jose, CA, USA) coupled with an HPLC method for the separation and quantitation of the ADP-ribosylated product as described previously [13]. PTx reference preparation (NIBSC, 90/518) was used as a standard. All other chemicals, unless specified otherwise, are of analytical grade and purchased from either Sigma-Aldrich or VWR-BDH (Poole, UK) as stated previously [13].
The PTx carbohydrate-binding activity in bacterial cell supernatant was performed as described by Gomez et al. [14]. Glycoproteins containing different N-glycans types were purchased from Sigma-Aldrich, UK: bovine pancreas ribonuclease B (RNaseB, R7884), foetal calf serum fetuin (Fetuin, F3004), foetal calf serum asialofetuin (AsFetuin, A4781), human plasma alpha-1 acid glycoprotein (AGP, G9885) and human transferrin (hTf, T3309) were used as carbohydrate ligands. Anti-PTx sheep serum (NIBSC Code: 97/572), monoclonal antibodies to S1 (NIBSC code 99/506), S2 and 3 (NIBSC code 99/534) and S4 (NIBSC code 99/554) subunits [33] were used as the detection antibody respectively. The relative potency of PTx binding activity in bacterial cell supernatant to PTx reference (90/518) was calculated using parallel line assay (CombiStats software, version 4.0, EDQM).
Animal Experiments
Ethics statement for animal
All animal experiments were approved by the Ethical Committee, National Institute for Biological Standards and Control, UK and regulated under the Animal Scientific Procedures Act (1986), UK.
Lymphocytosis test
All experiments were carried out using female NIH strain mice, aged approximately 3-4 weeks, obtained from Harlan, UK and maintained in pathogen-free conditions. Mice (10 per group) were injected intra-peritoneally (i.p.) with 0.5 ml of either PTx reference (NIBSC, 90/518) at 5.25, 1.75 and 0.58 IU/dose in phosphate buffered saline containing 0.2% (w/v) gelatine (PBSG) or bacterial strain supernatants with a PTx concentration of 100 ng/mouse. The negative control groups received PBSG only. At four days post administration, tail bleeds were taken from mice into a microfuge tube containing heparin solution (final heparin concentration of~10 units/ml). After mixing, leucocytes in the whole blood were counted using a Coulter counter (Coulter Ac.T 8 Series Analyser; Beckman Coulter, UK). The counts were expressed as the geometric mean of the number of lymphocytosis per µl of blood.
Dermal temperature histamine sensitisation test (HIST)
The histamine sensitisation test (HIST) was carried out as described previously by Ochiai [34]. In brief, mice (10 per group) were inoculated i.p. with 0.5 ml PBSG as negative control, PTx reference (90/518) at 5.25, 1.75 and 0.58 IU/dose in PBSG or with bacterial cell supernatant with a PTx concentration of 100 ng/mouse. Five days after inoculation, the mice were challenged i.p. with 0.5 ml of histamine solution (2 mg histamine base per dose) (Sigma, H7375). The dermal temperature of each mouse was measured 30 min after challenge using an infrared thermometer (IT-550, Horiba Ltd., Japan). The HIST activity in each bacterial cell supernatant was calculated in comparison to that of the reference toxin groups and expressed as IU/dose.
DNA sequencing
Strains were grown on charcoal agar for two days at 37°C. DNA was extracted using GenElute Bacterial Genomic DNA Kit (Sigma) according to the manufacturer’s instructions. The target genes were amplified by PCR using the following forward and reverse primers (Invitrogen) for each gene; ptxA: ptxA PCR F, CCCCCTGCC ATGGTGTGATC, ptxA PCR R, AGAGCGTCT TGCGGTCGATC; ptxB: ptxB PCR F, TGTACTACGA AAGCATCG, ptxB PCR R, TCACCAGCA CATAAGGAA; ptxC: ptxC PCR F, CTTCCGGAG GTTTCGACGTTTC, ptxC PCR R, TCTTTCAAGG GATTCATTCGC; ptxD: ptxD PCR F, GCTGCGGAAA ATGCTTTA; ptxD PCR R, TGGACAGTG AAGTTCTTG; ptxE: ptxE PCR F, GCCGTCTTC ATGCAACAAC, ptxE PCR R, AGGATGGG CAGAATGTGA. PCR was performed using a final volume of 50 µl which contained 10 µl 5X Green GoTaq Flexi Buffer, 4 µl 25 mM MgCl2, 1 µl dNTP mix, 0.25 µl Taq DNA Polymerase (all Promega), and 27.75 µl d H2O, 2 µl DNA sample, 2.5 µl of each primer pair at 10 µM. The following cycle was used: 1 cycle at 95°C for 2 min; step 2 was 30 cycles at 95°C for 30 s, 60°C for 30 s and 72°C for 30 s; the third step was 1 cycle at 72°C for 10 min. The products were analysed on 2% agarose gels (SeaKem) and purified from the gel using High Pure PCR Cleanup Micro Kit (Roche) according to the manufacturer’s instructions. Sequencing was performed using BigDye® Terminator v3.1 Cycle Sequencing Kit and ABI PRISM 3130xl Genetic Analyzer (Applied Biosytems) according to the manufacturer’s instructions. Briefly, a 10 µl amplification mix was prepared which contained 0.5 µM of the forward or reverse primer for the appropriate gene plus 4 µl purified H2O, 3 µl BigDye Terminator Ready Reaction Mix, 0.5 µl BigDye 5X Sequencing Buffer (all Applied Biosytems) and 1.5 µl DNA template. Amplification was performed with 30 cycles at 96°C for 30 s, 50°C for 15 s and 60°C for 30 s. Amplified DNA was precipitated by ethanol/EDTA according to the manufacturer’s instructions. The DNA was then re-suspended in 10 µl HiDi (applied biosytems), sequenced and sequencing data was analysed using Geneious 6.1.3.
Tandem mass spectrometric analysis
Purified PTx samples from strains PCRB12, PCRB20, B1865 and B2903 were diluted with 200 mM triethylammonium bicarbonate to bring the pH to 8.0 and denatured at 100°C for 5 min in the presence of RapiGest, an enzyme-compatible and acid-labile detergent (Waters, Milford, MA, USA). Once cooled, tryptic digestion was carried out at 37°C for 3h using manufacturer’s recommendation (Promega, Madison, USA) followed by adding HCl (0.45 N) to terminate the reaction and breakdown of RapiGest. The digests were desalted by the use of Pierce C18 spin columns following the manufacturer’s instruction (Product #89870PR; Thermo Scientific, Rockford, USA).
Mass spectrometric analysis was performed using a LTQ-Orbitrap Discovery mass spectrometer, coupled with an Ultimate 3000 nano-LC system (Thermo Fisher Scientific, UK). MS scan and MS/MS fragmentation were carried out in Orbitrap and LTQ respectively using top 5 data-dependent acquisition with dynamic exclusion mode enabled. The total cycle time was approximately 30 milliseconds. Data analysis including mass spectra processing and database searching were carried out using Thermo Proteome Discoverer 1.4 with built-in Sequest using UniProt FASTA database (released 2014_08). Initial mass tolerances by MS were set to 10 ppm. Up to two missed tryptic cleavages were considered and methionine oxidation was set as dynamic modification. All peptides at rank 1 with high confidence are considered to be unambiguously sequenced.
Statistical analysis
Student’s T-test was used to compare the difference of bioactivities between groups. A p-value ≤ 0.05 was considered as statistically significant.
Results
PTx ADP-ribosyltransferase and carbohydrate-binding activities in strains
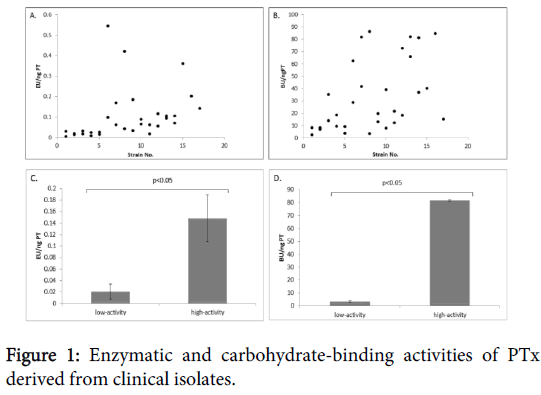
A total of 31 strains were examined for their PTx enzymatic and carbohydrate-binding activities. PTx ADP-ribosyltransferase activities were detected in all the strains and with a wide range from 0.005 to 0.545 EU/ng PTx (Figure 1). Of the six selected strains (3 low-and 3 high-activity), triplicate experiments of culture and respective assays were performed. The low-activity strains showed a range of enzymatic activities from 0.007-0.033 EU/ng PTx while the high-activity strains had a range of 0.120-0.195 EU/ng PTx (p<0.005) (Figure 1). Similarly, using an anti-PTx polyclonal antibody for detection, a wide range of carbohydrate-binding activities of PTx to glycoprotein fetuin was also observed (2.758-86.323 BU/ng PTx; Figure 1).
Results from triplicate experiments for the low-activity strains showed a range of 2.758 to 4.061 BU/ng PTx. The high-activity strains were approximately 20 fold higher binding activity (range of 81.193-82.262 BU/ng PTx) than the low-activity strains (p<0.005) (Figure 1).
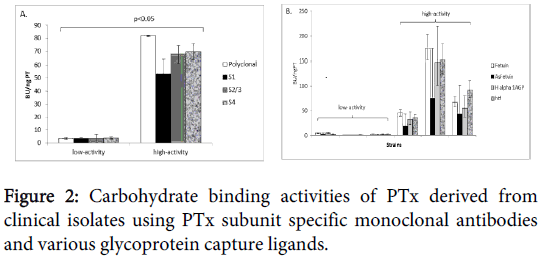
Monoclonal antibodies to different PTx subunits S1, S2 and 3 and S4 were also used as detection reagents to further investigate if the observed binding activities were strain specific (Figure 2).
In vivo toxicity activities in selected strains
The selected strains were also assessed for their in vivo toxicity by their ability to promote lymphocytosis. Mice injected with supernatants from the three high-activity strains showed higher lymphocyte counts compared to the low-activity strains although the difference was not significant (p=0.256; Figure 3). Further toxicity assessment, based on their ability to cause histamine sensitisation was carried out using the HIST model. All three supernatants from the high-activity strains showed significantly higher HS activities, by approximately 50%, than the low-activity strains (p=0.05; Figure 3).
DNA sequence and mass spectrometric analysis of PTx subunits
To further investigate the underlying mechanisms of the observed differences in the functional activities between these strains, genomic DNA sequencing and mass spectrometric analysis of PTx subunits were performed. The DNA sequences of the PTx gene clusters for all six strains were found to be identical (data not shown), except that there was a silent mutation of a single nucleotide change in ptxC (subunit S3) at base 681, where all the three low-activity strains contained cytosine in this location while all the three high-activity strains contained thymine (Table 2). This is a silent mutation as both codons code for the same amino acid cysteine (Cys199), the C-terminal residue of subunit S3.
| A. | ||||||||
|---|---|---|---|---|---|---|---|---|
| 676 | 679 | 682 | ||||||
| Consensus sequence Strains (type) | A T A | T G Y | T G A | |||||
| PRCB12 (ptxP1) | • • • | • • C | • • • | |||||
| PRCB20 (ptxP1) | • • • | • • C | • • • | |||||
| PRCB309 (ptxP1) | • • • | • • C | • • • | |||||
| PRCB414 (ptxP3) | • • • | • • T | • • • | |||||
| B1865 (ptxP3) | • • • | • • T | • • • | |||||
| B2903 (ptxP3) | • • • | • • T | • • • | |||||
| B. | ||||||||
| Strains | ||||||||
| PTx Subunit |
Peptide amino-acid sequence | PCRB12 (ptxP1) |
PCRB20 (ptxP1) |
B1865 (ptxP3) |
B2903 (ptxP3) |
|||
| S1 | SCQVGSSNSAFVSTSSSR (40-57) | + | + | - | - | |||
| S3 | VHVSKEEQYYDYEDATFQTALTGISLCNPA-ASIC (167-199) | + | + | - | ||||
Table 2: (A) DNA sequences of ptxC coding for subunit 3 from ptxP1 and ptxP3 strains at the single point mutation region (base 681) and (B) Mass spectrometric analysis of pertussis toxin (PTx) purified from ptxP1 and ptxP3 strains showing differences in tryptic peptides in subunits S1 and S3 that contain cysteine residues (underlined); (+) detected and (-) not detected.
Mass spectrometric analyses of tryptic peptides from subunits S1-S5 of PTx derived from four of these strains (two each of the low- and high-activity) were also carried out. Subunits S1 to S4 were clearly identified in all the PTx preparations and the amino acid sequence coverages compared favourably with published data [35,36]; for example, the data for strain PCRB12 were 86% for subunit S1, 35% for S2, 47% for S3, 53% for S4 while S5 was not detected. The tryptic peptides profiles of subunits S2 and S4 from all these strains were very similar (data not shown). However, there were differences amongst the cysteine containing tryptic peptides for subunits S1 and S3 between the high- and low-activity strains. In low-activity strains, cysteine containing tryptic peptides were identified between Cys41 and Cys201 regions of subunit S1 and at the C-terminal Cys199 region of subunit S3 (containing the silent mutation at base 681) (Table 2); and these peptides were not detected in the high-activity strains.
Discussion
B. pertussis is extremely homogenous and most variation observed so far has been in its virulence genes, for example: ptxA, pertactin and fimbriae [19,22]. In the present study, the strains tested had the same ptxA gene sequence. Since the 1990s the major PFGE group of B. pertussis circulating in Europe has been group IV [37,38] and this PFGE group IV is further divided into three subgroups, α,β and γ. In 2004, when isolates from eight European countries, including Finland and The Netherlands, were compared, isolates with subgroup IVβ were found to be dominant [37]. The ptxA gene codes for the A-protomer (subunit S1) of PTx which is an NAD hydrolase and an ADP-ribosyltransferase which catalyses the ADP-ribosylation of guanine nucleotide inhibitory (Gi) protein of eukaryotic cells. The B-oligomer (formed from subunit dimers S2-S4 and S3-S4 and subunit S5), which binds preferentially to multi-sialylated N-glycans structures [14], is responsible for the binding of the toxin to host cell receptors thereby facilitating entry of the A-protomer into the cell.
PTx is known to mediate bacterial attachment both to epithelial cells of the respiratory tract and to macrophages via its B-subunits [39,40]. In the present study, we measured the enzymatic and cell-binding function of the PTx [13,14] from clinically isolated strains and it was found that the high and low activities observed in the in vitro assays were mirrored the in vivo toxicity tests. This indicates a positive correlative relationship between the two in vitro assay methods and the in vivo methods. Furthermore, this in vitro biochemical assay system has been successfully used for the evaluation of PTx activities of different purified toxin preparations and residual toxin activities in vaccine matrices [11] and this is the first time it is applied to assessing the PTx toxic activities of clinical B. pertussis strains.
It has been reported that the B. pertussis population has changed after the introduction of vaccination [22]. One of these changes involves the promoter for PTx (ptxP) and strains carrying the ptxP1 allele haven been replaced by those carrying a novel variant, ptxP3. There is evidence that the ptxP3 strain-type is associated with increased incidences of pertussis [23,24]. There are certainly strong merits to further study these two pertussis strain types in terms of their in vitro activities, genetic make-ups and the relationship of PTx structure to bioactivities. In the present study, a total of 31 clinical isolated strains (17 ptxP1 strains and 14 ptxP3 strains) were assessed for PTx enzymatic and carbohydrate binding activities and a wide range of activities were found for both assay systems. Interestingly the three strains with highest in vitro activities belonged to the ptxP3 type and the three strains with the lowest activities belonged to the ptxP1 type. However, not all ptxP3 strains showed higher activity than ptxP1 strains and there was no statistical significant difference between these two groups with all strains tested (data not shown). Therefore, the present study was unable to establish a clear difference between these two types of strains in terms of their enzymatic and carbohydrate binding activities. Future study involving larger number of ptxP1 and ptxP3 strains would provide a better definitive analysis.
The positive relationship between the two biochemical functional activities and the in vivo toxic activities in those clinical isolates is interesting. One could speculate that the significantly higher carbohydrate binding activity to various N-glycans structures of glycoproteins by the high-activity strains (type ptxP3) found in this study may explain their increased colonising ability and toxicity in relation to the low-activity strains (type ptxP1) by enhanced host cell binding activities [41]. Genotype analysis does not directly account for differences in specific activities between the two strain types, as the DNA coding for PTx of all strains are the same, except for the silent point mutation in ptxC (subunit S3) at base 681 in high-activity strains relative to low-activity ones. The three low-activity strains contained cytosine at this location, whereas high-activity strains contained thymine and this finding is in agreement with a previous report [23]. It has been suggested that naturally occurring silent single nucleotide polymorphisms (SNPs) can affect in vivo protein folding and consequently affect protein function via altered translational kinetics of mRNA, which in turn might affect final protein conformation [42] and therefore give rise to a protein product with the same amino acid sequence but different structural or functional features [43,44]. Another explanation for the different specific activities may be due to differences in post-translational modification of PTx subunits in low- and high-activity strains. This may explain why mass spectrometric analysis showed differences in the peptides detected when PTx from the low- and high-activity strains were compared. Under non-reducing conditions, tryptic peptides with free cysteine residues were detected in subunits S1 and S3 from the low-activity strains which would indicate disulphide bonds were not formed in those peptide regions. In contrast, those same peptides were not detected from the high-activity strains, which would suggest disulphide bonding amongst those cysteine residues. Absence of a peptide might also indicate post-translational modification. Genetic loci, located outside the PTx gene cluster may be involved in post-translational modification. In this context it is significant that the genomes of low- and high-activity strains differ in 31 genes of which, four are of unknown functions [45]. In future, more detailed proteomic studies are required to understand, and to explore any differences in post-translational modifications between the two types of strains, and how these affect the enzymatic and carbohydrate binding activities. Preferably, a larger number of clinical isolates from different countries should be examined.
In conclusion, in the present study, we report for the first time that PTx produced by clinically isolated strains showed differences in their enzymatic and carbohydrate binding activities. The positive relationship between the in vitro and in vivo assays indicate that the biochemical assay results reflect in vivo toxicities and provides evidence that the in vitro assay system could be applied to the evaluation of clinical isolates. It is also interesting to note that the difference between low- and high-activity strains could be related to differences in PTx structure and/or due to post-translational modification. These findings should give impetus for further studies on the subtle differences between strains producing this key virulence factor of B. pertussis and vaccine antigen critical to whooping cough disease prevention.
Acknowledgements
The authors thank Dr Ed Mee for his help with the DNA sequence work. We are also grateful for the support provided by the Biological Services Division at NIBSC.
References
- Munoz JJ (1985) Biological activities of pertussigen (pertussis toxin). In: Sekura RD, Moss J, Vaughan M (eds.) Pertussis toxin. Academic Press Inc., London, pp1-18.
- Miller D, Madge N, Diamond J, Wadsworth J, Ross E (1993) Pertussis immunisation and serious acute neurological illnesses in children. BMJ 307: 1171-1176.
- Donnelly S, Loscher CE, Lynch MA and Mills KHG (2001) Whole-cell but not acellular pertussis vaccines induce convulsive activity in mice: evidence of a role for toxin-induced interleukin-1beta in a new murine model for analysis of neuronal side effects of vaccination. Infect Immun 69: 4217-4223.
- Yamamoto A, Nagata N, Ochiai M, Kataoka M, Toyoizumi H, et al. (2002) Enhanced sensitisation of mice with diphtheria tetanus acellular pertussis vaccine to local swelling reaction to the booster immunisation. Vaccine 20: 3088-3094.
- Pierce C, Klein N, Peters M (2000) Is leukocytosis a predictor of mortality in severe pertussis infection? Intensive Care Med 26: 1512-1514.
- Guidelines for the production and control of the acellular pertussis component of monovalent or combined vaccines (1998) In: WHO Technical Report Series 878, 57-72.
- Pertussis vaccine (acellular, component, adsorbed). In: European Pharmacopoeia, version 7.0, 01/2008:1356: 806-807.
- Locht C, Keith JM (1986) Pertussis toxin gene: nucleotide sequence and genetic organization. Science 232: 1258-1264.
- Tamura M, Nogimori K, Murai S, Yajima M, Ito K, et al. (1982) Subunit structure of islet-activating protein, pertussis toxin, in conformity with the A-B model. Biochemistry 21: 5516-5522.
- Ui M (1990) ADP-ribosylating toxins and G protein (Moss J and Vaughan M Editors), pp 45-77. American Society for Microbiology, Washington, D.C.
- Yuen C-T, Horiuchi Y, Asokanathan C, Cook S, Douglas-Bardsley A, et al. (2010) An in vitro assay system as a potential replacement for the histamine sensitisation test for acellular pertussis based combination vaccines. Vaccine 28: 3714-3721.
- Cyr T, Menzies AJ, Calver J and Whitehouse LW (2001) A quantitative analysis for the ADP-ribosylation activity of pertussis toxin: an enzymatic-HPLC coupled assay applicable to formulated whole cell and acellular pertussis vaccine products. Biologicals 29: 81-95
- Yuen CT, Canthaboo C, Menzies JA, Cyr T, Whitehouse LW, et al. (2002) Detection of residual pertussis toxin in vaccines using a modified ribosylation assay. Vaccine 21: 44-52.
- Gomez SR, Xing DK, Corbel MJ, Coote J, Parton R, et al. (2006) Development of a carbohydrate binding assay for the B-oligomer of pertussis toxin and toxoid. Anal Biochem 356: 244-253.
- Tan Y, Fleck RA, Asokanathan C, Yuen CT, Xing D, et al. (2013) Confocal microscopy study of pertussis toxin and toxoids on CHO-cells. Hum VaccinImmunother 9: 332-338.
- Ochiai M, Horiuchi Y, Yuen C-T, Asokanathan C, Yamamoto A, et al. (2014) Investigation in a murine model of possible mechanisms of enhanced local reactions to post-primary diphtheria-tetanus toxoid boosters in recipients of acellular pertussis diphtheria-tetanus vaccine. Human Vaccines &Immunotherapeutics 10: 2074-2080
- Guiso N (2014) Pertussis vaccination and whooping cough: and now what? Expert Rev Vaccines 13: 1163-1165.
- He Q, Mertsola J (2008) Factors contributing to pertussis resurgence. Future Microbiol 3: 329-339.
- Mooi FR (2010) Bordetella pertussis and vaccination: the persistence of a genetically monomorphic pathogen. Infect Genet Evol 10: 36-49.
- Guiso N, Wirsing von Konig C, Forsyth K, Tan T,Plotkin SA (2011) The global pertussis initiative: report from a round table meeting to discuss the epidemiology and detection of pertussis, Paris, France, 11-12 January 2010. Vaccine 29: 1115-1121.
- van Boven M, Mooi FR, Schellekens JF, de Melker HE, Kretzschmar M (2005) Pathogen adaptation under imperfect vaccination: implications for pertussis. ProcBiolSci 272: 1617-1624.
- Kallonen T, He Q (2009) Bordetella pertussis strain variation and evolution postvaccination. Expert Rev Vaccines 8: 863-875.
- Mooi FR, van Loo IH, van Gent M, He Q, Bart MJ, et al. (2009) Bordetella pertussis strains with increased toxin production associated with pertussis resurgence. Emerg Infect Dis 15: 1206-1213.
- Octavia S, Sintchenko V, Gilbert GL, Lawrence A, Keil AD, et al. (2012) Newly Emerging Clones of Bordetella pertussis Carrying prn2 and ptxP3 Alleles Implicated in Australian Pertussis Epidemic in 2008-2010. J Infect Dis 205: 1220-1224.
- Advani A, Gustafsson L, Ahrén C, Mooi FR, Hallander HO (2011) Appearance of Fim3 and ptxP3-Bordetella pertussis strains, in two regions of Sweden with different vaccination programs. Vaccine 29: 3438-3442.
- Kallonen T, Mertsola J, Mooi FR, He Q (2012) Rapid detection of the recently emerged Bordetella pertussis strains with the ptxP3 pertussis toxin promoter allele by real-time PCR. ClinMicrobiol Infect 18: E377-379.
- Caro V, Elomaa A, Brun D, Mertsola J, He Q, et al. (2006) Bordetella pertussis, Finland and France. Emerg Infect Dis 12: 987-989.
- Sato Y, Cowell JL, Sato H, Burstyn DG, Manclark CR (1983) Separation and purification of the hemagglutinins from Bordetella pertussis. Infect Immun 41: 313-320.
- Pourahmadi A, Esmaili F, Zavaran H-A, Rezaee A, Salehi E, et al. (2004) Proliferative response to purified Bordetella pertussis toxin on murine spleen lymphocytes. Arch Razi Ins 58:63-71.
- Xing D, Das RG, Newland P and Corbel M (2002) Comparison of the bioactivity of reference preparations for assaying Bordetella pertussis toxin activity in vaccines by the histamine sensitisation and Chinese hamster ovary-cell tests: assessment of validity of expression of activity in terms of protein concentration. Vaccine 20: 3535-3542.
- Stott DI (2000) Immunoblotting, dot-blotting, and ELISPOT assays: methods and applications. J Immunoassay 21: 273-296.
- Sato H, Sato Y (1994) Structural relationship between the S1 and S4 subunits of pertussis toxin. FEMS MicrobiolLett 115: 63-69.
- Sato H, Sato Y (1990) Protective activities in mice of monoclonal antibodies against pertussis toxin. Infect Immun 58: 3369-3374.
- Ochiai M, Yamamoto A, Kataoka M, Toyoizumi H, Arakawa Y et al. (2007) Highly sensitive histamine-sensitization test for residual activity of pertussis toxin in acellular pertussis vaccine. Biologicals 35: 259-264.
- Tummala M, Lee SM, Chess E, Hu P (2010) Characterization of pertussis toxoid by two-dimensional liquid chromatography-tandem mass spectrometry. Anal Biochem 401: 295-302.
- Williamson YM, Moura H, Schieltz D, Rees J, Woolfitt AR, et al. (2010) Mass spectrometric analysis of multiple pertussis toxins and toxoids. J Biomed Biotechnol 2010: 942365.
- Caro V, Njamkepo E, Van Amersfoorth SC, Mooi FR, Advani A, et al. (2005) Pulsed-field gel electrophoresis analysis of Bordetella pertussis populations in various European countries with different vaccine policies. Microbes and Infection 7: 976-982.
- Hallander H, Advani A, Riffelmann M, von König CH, Caro V, et al. (2007) Bordetella pertussis strains circulating in Europe in 1999 to 2004 as determined by pulsed-field gel electrophoresis. J ClinMicrobiol 45: 3257-3262.
- Menozzi FD, Debrie AS, Tissier JP, Locht C, Pethe K, et al. (2002) Interaction of human Tamm-Horsfall glycoprotein with Bordetella pertussis toxin. Microbiology 148: 1193-1201.
- van'tWout J, Burnette WN, Mar VL, Rozdzinski E, Wright SD, et al. (1992) Role of carbohydrate recognition domains of pertussis toxin in adherence of Bordetella pertussis to human macrophages. Infect Immun 60: 3303-3308.
- van Loo IHM (2012) Polymorphism in the promoter region of pertussis toxin: Has vaccination selected for more virulent strains? In: Vaccine-driven evolution of Bordetella pertussis: Changes in population structure and strain fitness, Chapter 6, PhD Thesis, Universiteity Utrecht,
- Komar AA, Lesnik T, Reiss C (1999) Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett 462: 387-391.
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, et al. (2007) A "Silent" Polymorphism in the MDR1 Gene Changes Substrate Specificity. Science 315: 525-528.
- Komar AA (2007) Genetics. SNPs, silent but not invisible. Science 315: 466-467.
- King AJ, van Gorkom T, Pennings JL, van der Heide HG, He Q, et al. (2008) Comparative genomic profiling of Dutch clinical Bordetella pertussis isolates using DNA microarrays: identification of genes absent from epidemic strains. BMCGenomics 9:311.
Citation: Asokanathan C, Yuen CT, Kmiec D, Wheeler JX, Markey K et al. (2015) Functional Differences in Pertussis Toxins from Bordetella pertussis Clinical Isolates as Determined by in vitro and in vivo Assays. J Clin Exp Pathol 5:243. DOI: 10.4172/2161-0681.1000243
Copyright: © 2015 Asokanathan C et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 14833
- [From(publication date): 10-2015 - Nov 22, 2024]
- Breakdown by view type
- HTML page views: 10368
- PDF downloads: 4465