Fast Twitch Skeletal Muscle Remodeling by Prolonged Endurance Exercise is Associated with Crosstalk between Anabolic and Catabolic Signaling Pathways in Mice
Received: 22-Jan-2018 / Accepted Date: 07-Mar-2018 / Published Date: 15-Mar-2018 DOI: 10.4172/2168-9652.1000228
Abstract
Endurance exercise (EXE) is a potent inducer of both muscle fiber transformation as well as autophagy in skeletal muscles. However, it has remained unknown whether autophagy is associated with EXE-induced muscle fiber transformation. Thus, we examined autophagy responses in in fast muscle (tibialis anterior; TA) tissues after sixweek long term treadmill EXE by assessing a series of autophagy signaling pathways and muscle fiber transformation through Western blot analysis and fluorescence microscopy. First, we confirmed that EXE caused slow muscle phenotypes in TA muscle, evidenced by reduction in cross-sectional areas of muscle fibers, increases in type I and II fibers, and upregulation of mitochondrial proteins. Subsequently, our results showed that transformation of muscle fiber types concurred with autophagy upregulation (e.g., an increase in LC3-II, LC3-II/I ratio, and BNIP3); however, intriguingly, inductive signaling of autophagy (e.g., phosphorylation of AMPK and BCL2) were suppressed. Moreover, anabolic signaling (e.g., an increase in phosphorylation levels of AKT, mTOR, p70S6K, and FOXO3), which typically serves as anti-autophagy factor were significantly elevated. Our findings suggest that although autophagy levels were sustained higher in TA of EXE-trained mice compared to sedentary mice, concomitant potentiation of anabolic signaling by EXE may serve as a negative feedback to prevent excessive catabolism induced by autophagy. We also suggest that this anabolic response may be necessary for remodeling of anaerobic fast muscle fibers into aerobic muscle fibers and mitochondrial biogenesis.
Keywords: Endurance exercise; Autophagy; Mitochondria; Skeletal muscle; Anabolic signalling; Muscle fiber type
Introduction
Skeletal muscle is a highly dynamic, plastic tissue and thus quickly responds to cellular stimuli such as mechanical stress caused by physical exercise. Chronic endurance exercise has been known to modulate morphological (e.g., reduction in cross-sectional area) and biochemical (e.g., muscle fiber-type shifts from fast muscle fibers to slow muscle fibers and mitochondrial biogenesis) changes. These changes contribute to improvement of muscular endurance and metabolic efficiency [1-6]. A recent study has shown that endurance exercise (EXE)-induced biophysical adaptations in skeletal muscle is associated with protein turnover: the balance between muscle protein synthesis and muscle protein breakdown [7]. Given a study showing that prolonged transition time of intracellular calcium during endurance muscle contraction and relaxation plays an important role in mediating aerobic phenotypes in fast muscle tissues [8,9], calciummediated signaling has long been considered as a potential mechanism associated with EXE-mediated muscular adaptation. For example, increased calcium transition during EXE allows calcium-activated protease CALPAIN to cleave structural and contractile proteins, which are removed by an ubiquitin-proteasome system (UPS), resulting in downsizing muscle mass [10]. Aside from the UPS in association with induction of aerobic phenotypes of skeletal muscle [11], autophagy has emerged as a new potential regulator of protein turnover [12].
Autophagy is a catabolic process involved in removal of cellular cargos such proteins, lipids, and small organelles, which requires the several sequential signaling cascades, including initiation of the isolated membrane, nucleation, elongation, maturation, and degradation [13,14]. Autophagy levels are generally determined by changes in the amount of lipidated form of microtubule-associated protein light chain 3-II (LC3-II) as well as p62 and BCL2/adenovirus E1B 19kDA protein-interacting protein 3 (BNIP3) and other ATG proteins including BECLIN-1, ATG7. To terminate the whole autophagy process, the consecutive interplay between autophagosomes and lysosomes via lysosome-associated membrane protein and CATHEPSINS are required [15,16]. Recent studies have provided strong evidence that EXE induces autophagy in skeletal muscle tissues [17] and play a potential role in skeletal muscle homeostasis [18]. However, the functional role of autophagy in transformation of muscle fiber types and metabolic adaptation in fast muscle tissues in response to EXE remains poorly understood.
Although autophagy plays an important role in maintaining muscular homeostasis, excessive autophagy can result in muscular atrophy and degeneration [19]. Thus, proper operation of autophagy with precise orchestration of numerous molecular interactions is critical. Adenosine monophosphate (AMP)-activated kinase (AMPK), a master regulator of metabolism in many tissues including skeletal muscle is a critical regulator of autophagy under numerous cellular stress conditions [20-25]. Activated AMPK phosphorylates its downstream signaling molecule ULK1 at Ser555 and thus potentiates a class III PI3 kinase to induce autophagy. By contrast, activation of a mammalian target of rapamycin (mTOR) via anabolic signaling cascade by AKT antagonizes the autophagic process by phosphorylating ULK1 at a different phosphorylation site (Ser757) [26]. Thus, modulation of AMPK and mTOR activation status determines autophagy initiation and abolition. Although a recent study has reported that EXE-induced autophagy is correlated with transformation of muscle phenotypes from fast twitch fiber to slow twitch fibers [27], complex signaling nexus of EXE-induced autophagy pathways (AMPK vs . mTOR) and its association with muscle fiber type shifting in fast muscle tissues remain to be elucidated.
In this study, we sought to examine potential relationships between endurance exercise-induced autophagy and muscular phenotype changes in tibialis anterior (TA) muscle and investigate a potential adaptive mechanism of autophagy regulation in response to long-term EXE. Our study showed that EXE-induced autophagy occurs despite the disrupted induction process of autophagy and is associated with aerobic phenotypes in TA muscle. Furthermore, the present study provides potential evidence that enhanced anabolic signaling cascade may be necessary for prohibiting excessive activation of autophagy but for promoting protein synthesis for the conversion of muscle fiber types and mitochondrial biogenesis.
Materials and methods
Animals
Male C57BL/6 mice (N=28, 8 weeks old) were purchased from ENVIGO company (Tampa, FL, USA) and housed in a temperature (22˚C) and humidity (55%)-controlled room under a 12 h light:12 h dark cycle with a standard chow diet and water ad libitum. All experiments were conducted according to the Declaration of Helsinki regulations addressing the welfare of experimental animals and were approved by the Institutional Animal Care and Use Committee of the University of West Florida (#2015-002). After one-week of acclimation period, the animals were divided into two groups: sedentary control group (CON, n=14) and endurance exercise group (EXE, n=14).
Endurance exercise protocol
Endurance exercise was performed using a modified motor-driven animal treadmill. The animals assigned to EXE were allowed to be familiarized with treadmill running with slow speed (at 9 meter/min for 30 min), and then the speed was gradually increased up to 15 meter/min throughout the one-week acclimation period. After the adaptation, the animals assigned to the EXE group performed treadmill running exercise at 15 meter/min at 0% incline for 60 min per day, 5 days per week for 6 weeks. To preclude possible confounding results that may occur due to electrical shocks, we did not use electrical grids but instead applied soft plastic brushes at the end of each lane. Animals touching the brush become aroused and continued running. We followed our animal use protocol that any animals failing to run despite continuous touch with brushes would be allowed to terminate their exercise to avoid unwanted stress responses. In our study, all animals successfully completed the exercise protocol. Animals assigned to sedentary control group were exposed to the same degree of environmental stress (e.g., noise from treadmill).
Tissue extraction
The animals were sacrificed by cervical dislocation, and tibialis anterior (TA) muscle was excised right after 90 min of the last session of endurance exercise training to examine the peak autophagy response according to recent studies [12,23]. We chose TA muscle because this muscle consists of mostly fast-twitch fibers (type IIb and IIa) without slow-twitch fiber (type I) in C57BL/6 mice [28], such that adaptive responses of fast muscle fibers to endurance exercise would be optimally observed. Five animals were used for tissue histology and nine animals for biochemical assays for each group. Once animals reached a surgical plane of anesthesia, the TA muscles were excised quickly. Muscle tissues assigned for the histological analysis were embedded with OCT freezing medium and frozen by submerging it in isopentane chilled in liquid nitrogen. Other tissues were stored in cryogenic storage tubes and kept at -80˚C until needed for Western blot analysis.
Skeletal muscle fiber-type composition and cross-sectional area
Frozen TA muscles were cross-sectioned at 10 μm in the middle section with a cryostat microtome (LEICA CM 1860, Germany), and the sectioned samples were collected on glass slides. The tissue samples were air-dried for 30 min at room temperature (RT) prior to histological processing. Then, they were permeabilized with 0.5% Triton X-100 mixed with phosphate-buffered saline (PBS) for 30 min at RT, followed by 5 min of washing with PBS. After the wash, each tissue sample was delimited in a circle drawn using ImmEdge™ pen (H-4000, Vector Laboratories, USA). The tissues were blocked with 5% normal goat serum blocking solution (50062Z, ThermoFisher Scientific, USA) for 1 h at RT, followed by wash (3 × 5 min) with PBS. Thereafter, two different Fab blockers (AffiniPure Fab Fragment Goat Anti-Mouse IgG, #115-007-003, Jackson Immnoresearch, USA and AffiniPure Fab Fragment Goat Anti-Mouse IgM, μ Chain Specific, #115-007-020, Jackson Immnoresearch, USA) were applied to the tissues to eliminate endogenous IgG and IgM cross-activity. After 1 h incubation at RT, the tissue sections were washed (3 × 5 min) with PBS, and designated primary antibodies, dystrophin (1:100, #PA1-21011, ThermoFisher, USA), myosin heavy chain type I (1:10, #A4.840, Developmental Studies Hybridoma Bank, USA), and myosin heavy chain type IIa (1:25, #SC-71, Developmental Studies Hybridoma Bank, USA) were incubated for 1 h at RT. The tissue sections were then washed with PBS (3 × 5 min), and secondary antibodies (Alexa Fluor 488, 1:50, #111-545-144, Jackson ImmunoResearch, USA; AMCA 350, 1:50, #115-155-020, Jackson ImmunoResearch); and Cy™ 3, 1:50, #115-165-071, Jackson ImmunoResearch) were applied and incubated for 1 h at RT under dark environment. After washing (3 × 5 min) with PBS, the tissue sections were mounted with VECTASHIELD®Hard +Set™ Mounting Medium (H-1400, Vector Laboratories, USA) under cover glasses, and sealed with a nail polisher. The tissues’ fluorescence images were visualized and captured using EVOS Auto inverted fluorescence microscope (Life Technologies™, #AMAFD1000) provided with three filters (DAPI, GFP, and RFP).
An analysis of cross-sectional area (CSA) of muscle fibers was conducted in accordance with published references [27]. Briefly, the values of CSA were obtained by measuring circumferences (greencolored dystrophin) of total 60 random muscle fibers acquired from five random fields, using Image J software (NIH, USA). Also, the composition of myosin heavy chain (MHC) isoforms were assessed by counting the number of each muscle fiber: type I (blue), type IIa (red), and type IIx+b (dark).
Detection of autophagy positive muscle fibers
Determination of autophagy-positive muscle fibers was based upon counting the number of fibers displaying accumulation of green fluorescence-tagged LC3 puncta in TA muscle tissues using immunofluorescence microscopy. Muscle tissues were cross-sectioned (10 μm thick) using a cryostat and were fixed with 4% formaldehyde made with PBS (pH 7.4) on the ice for 20 min. The cross-sectioned tissues were washed for 10 min with PBS and permeabilized in 0.3% triton X-100 with 10 mM sodium citrate solution (pH=6.0) for 20 min at room temperature. After being washed with PBS for 10 min, the tissues were blocked with 5% normal goat-serum for 30 min at room temperature and incubated with LC3 antibodies (Cell signaling, #4108, 1:200) prepared in 5% normal goat-serum in PBS at 4°C overnight. Next day, the tissues were washed with PBS for 15 min and incubated with goat anti-rabbit Alexa Fluor®488 (Jackson ImmunoResearch, #111-545-144, 1:200) for 30 min at RT. The tissues were washed for 10 min with PBS and incubated with NucBlue™ Live Cell Stain Ready Probes™ reagent (Lifetechnologies, #R10477) for 2 min at RT. The tissues were washed for 10 min with PBS and mounted with VECTASHIELD® Mounting medium solution (VECTOR, #H-1400). Green fluorescence puncta were probed using EVOS Auto fluorescence microscope (Life Technologies™, #AMAFD1000). The percentage of LC3-positive puncta in muscle fibers was calculated.
Western blot analysis
TA muscles were homogenized 1:20 w/v in T-PER® buffer (Thermo Scientific, #78510, Waltham, MA) containing a Halt™ Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific, #78446, Waltham, MA), using a Polytron™ PT 2500E Homogenizer (Kinematica, #08-451-320, Bohemia, NY). Tissue homogenates were centrifuged at 10,000 × g for 15 min for tissue protein extraction. Protein concentration was assessed using a Pierce™ Coomassie Plus Assay Kit (Thermo Scientific, #23236, Waltham, MA). For Western blot assays, 50 μg of proteins were separated by SDS-PAGE using Bolt™ 4-12% Bis- Tris Plus Gels (Invitrogen, #NW04125BOX, Carlsbad, CA) for 1 h at 80 volt and then transferred to PVDF membranes for 1 h at 30 volt in a cold condition. Non-specific proteins were blocked for 60 min at room temperature in a blocking solution (5% BSA in Tris-buffered saline containing 0.1% tween-20), and then the membranes were incubated with designated primary antibodies overnight at 4α°C. The antibodies used are as follows: LC3 (#4108, 1:1000), p62 (#5114, 1:1000), BNIP3 (#3769, 1:1000), AMPKα (#2532, 1:1000), p-AMPKαThr172 (#2535, 1:1000), BECLIN-1 (#3738, 1:1000), ATG7 (#2631, 1:1000), AKT (#9272, 1:1000), p-AKTSer473 (#9271, 1:1000), mTOR (#2972, 1:1000), p-mTORSer2448 (#2971, 1:1000), ULK1 (#8054, 1:1000), p-ULKSer555 (#5869, 1:1000), and p-ULK1Ser757 (#14202, 1:1000) from Cell Signaling Technology; LAMP-2 (#PA1-655, 1:1000), DRP1 (#PA1-16987, 1:500), p-DRP1Ser637 (#PA5-37534, 1:500), and OPA1 (#MA5-16149, 1:1000) from ThermoFisher Scientific; CATHEPSIN L (#133641, 1:500), MitoProfile® Total OXPHOS (#110413, 1:1000), MFN2 (#56889, 1:200), and NRF-1(#175932, 1:1000) from Abcam; BCL2 (#7382, 1:500), p-BCL2Ser87 (#377576, 1:500), p70S6K (#8418, 1:500), p-p70S6KSer411 (#8416, 1:500), and TFAM(#376672, 1:500) from Santa Cruz Biotechnology; FOXO3α (#NBP2-16521, 1:1000), p- FOXO3αSer253 (#NBP1-19927, 1:1000), and PGC-1α (#NBP1-04676, 1:1000) from Novus Biologicals. After the overnight incubation, the membranes were washed three times of 10 min with 1X TBS-T and then incubated with designated secondary antibodies conjugated with HRP for 1 h at room temperature: goat anti-rabbit (#1148960, 1:4000), rabbit anti-goat (#811620, 1:2000) from ThermoFisher Scientific; goat anti-mouse (#2005, 1:5000) from Santa Cruz Biotechnology. Then, the membranes were washed 3 times of 10 min in 1X TBS-T, after which immunoreactivity was detected with SuperSignal™ West Dura Extended Duration Substrate (Thermo Scientific, #34076, Waltham, MA). The digital images were acquired using ChemiDoc™ MP Imaging System (Bio-Rad, Hercules, CA), and band intensities were quantified by using Image Lab™ software version 5.2.1 (Bio-Rad, Hercules, CA). The digital intensity of all targeted protein images was normalized by the intensity of Ponceau (#P7170, Sigma-Aldrich, St. Louis, MO)-stained proteins on the same membrane to verify equal loading and quality of transfer.
Statistical analysis
All data were analyzed using the SPSS 22.0 software package (SPSS, Chicago, IL, USA). Values in the table and bar graphs were expressed as means ± SEM and fold changes, respectively. Independent, unpaired t-test was performed to examine the significant difference between two groups (control vs. exercise group). A statistical significance level was established at p<0.05.
Results
Endurance exercise elicits slow muscle phenotypes in TA muscles
To examine whether long-term endurance exercise (EXE) induces aerobic morphological adaptation in TA muscle tissues, a ratio of TA muscle mass to body weight and muscle mass (wet weight) were measured. As presented in (Table 1). TA muscle weight was significantly decreased in EXE (93.9 ± 1.61 g) group compared with CON group (101.7 ± 3.06 g), without significant changes in body weight and relative muscle mass between groups.
| CON | EXE | P | |
|---|---|---|---|
| Body weight (g) | 29.6 ± 0.87 | 27.9 ± 0.74 | 0.079 |
| TA muscle mass (mg) | 101.7 ± 3.06 | 93.9 ± 1.61 | 0.021* |
| TA muscle mass/ body weight (mg/g) | 3.46 ± 0.14 | 3.38 ± 0.09 | 0.329 |
Table 1: Effects of six weeks of endurance exercise in changes of body weight, muscle mass of TA, and a ratio of muscle mass of TA to body weight. Values are expressed as a mean ± SEM. * indicates significant difference compared to CON at p<0.05.
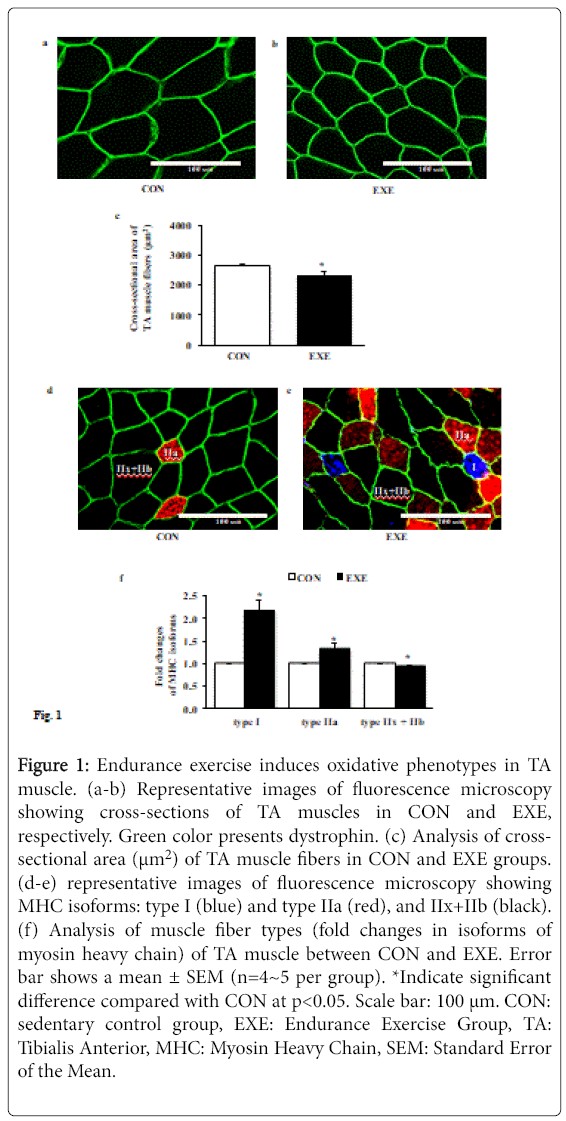
Next, we assessed whether EXE alters the cross-sectional area (CSA) of individual muscle fiber and induces a shift of fiber types. Data from immunofluorescence microscopy showed that the CSA of TA muscle fibers after EXE was significantly diminished compared with CON group (Figures 1a-1c). In addition, EXE acquired highly oxidative slow muscle fibers (type I, blue), which was not observed at all in a control group and increased the number of fatigue resistant oxidative intermediate myofibers (type IIa, red), whereas the number of highly glycolytic fast myofibers (type IIx and IIb, black) was significantly reduced (Figures 1d-1f).
Figure 1: Endurance exercise induces oxidative phenotypes in TA muscle. (a-b) Representative images of fluorescence microscopy showing cross-sections of TA muscles in CON and EXE, respectively. Green color presents dystrophin. (c) Analysis of crosssectional area (μm2) of TA muscle fibers in CON and EXE groups. (d-e) representative images of fluorescence microscopy showing MHC isoforms: type I (blue) and type IIa (red), and IIx+IIb (black). (f) Analysis of muscle fiber types (fold changes in isoforms of myosin heavy chain) of TA muscle between CON and EXE. Error bar shows a mean ± SEM (n=4~5 per group). *Indicate significant difference compared with CON at p<0.05. Scale bar: 100 μm. CON: sedentary control group, EXE: Endurance Exercise Group, TA: Tibialis Anterior, MHC: Myosin Heavy Chain, SEM: Standard Error of the Mean.
Endurance exercise sustains high levels of autophagy in TA muscle despite attenuated AMPK activities
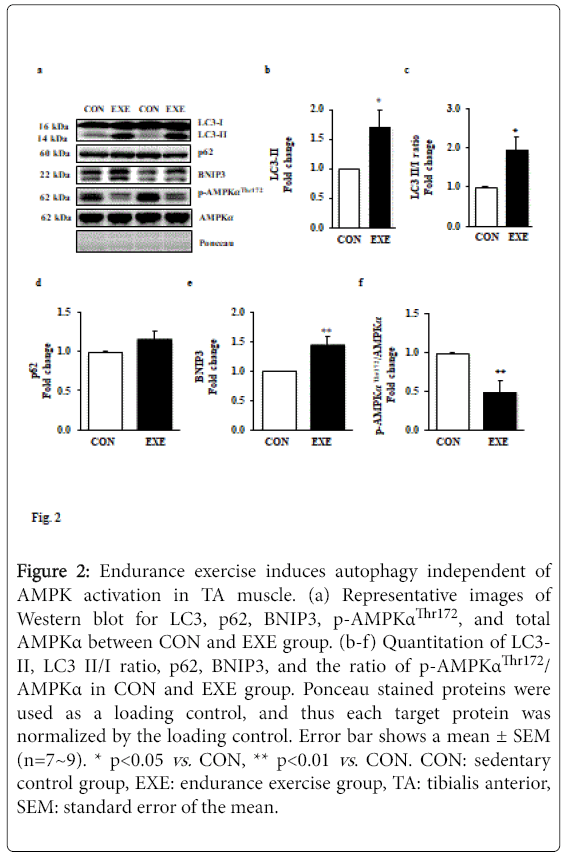
Since we observed a significant reduction in muscle mass, and autophagy is a potential catabolic process, we explored a question of whether exercise-induced autophagy is associated with the reduced fiber cross-sectional area. We assessed a key autophagy index protein LC3-II and found that EXE significantly increased LC3-II (Figures 2a and 2b) as well as the LC3-II to I ratio (Figures 2a and 2c) in EXE group compared to CON group. To confirm if the increase in LC3-II is due to an enhanced autophagy flux, we next measured levels of p62 since it has been reported to be a marker for autophagy flux. Interestingly, p62 levels were not changed (Figures 2a and 2d) but a potent autophagy inducer, BNIP3, was significantly elevated in the EXE group, compared with CON group (Figures 2a and 2e).
Figure 2: Endurance exercise induces autophagy independent of AMPK activation in TA muscle. (a) Representative images of Western blot for LC3, p62, BNIP3, p-AMPKαThr172, and total AMPKα between CON and EXE group. (b-f) Quantitation of LC3- II, LC3 II/I ratio, p62, BNIP3, and the ratio of p-AMPKαThr172/ AMPKα in CON and EXE group. Ponceau stained proteins were used as a loading control, and thus each target protein was normalized by the loading control. Error bar shows a mean ± SEM (n=7~9). * p<0.05 vs. CON, ** p<0.01 vs . CON. CON: sedentary control group, EXE: endurance exercise group, TA: tibialis anterior, SEM: standard error of the mean.
Given that AMPK activity has been suggested to regulate induction of autophagy, we next measured the levels of phosphorylated AMPK. Our data showed that phosphorylation levels of AMPK were remarkably suppressed in response to exercise compared with those in CON group (Figures 2d and 2f).
Endurance exercise interferes in autophagy induction processes without modulating lysosomal proteins in TA muscle
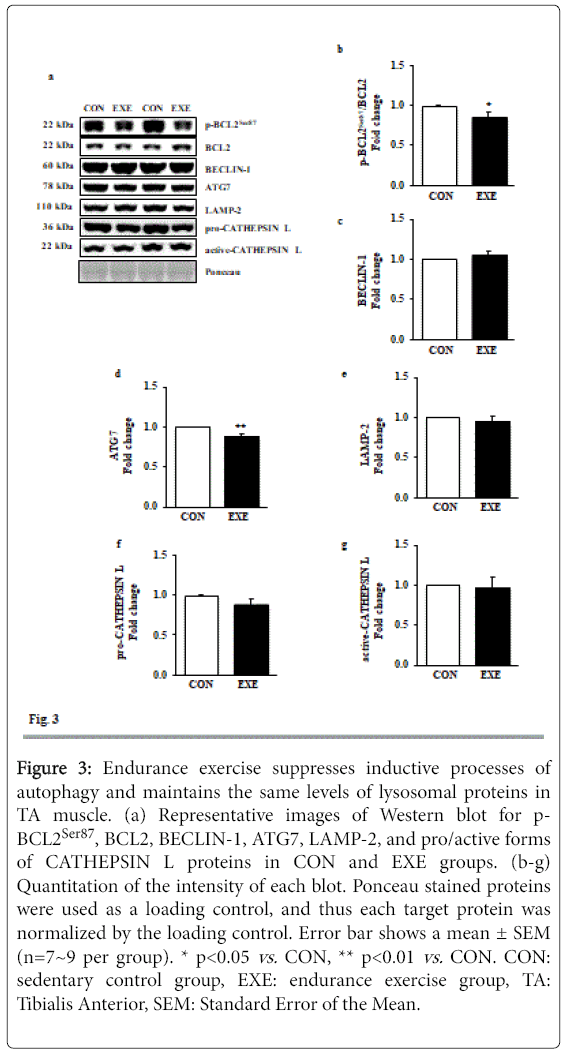
Since dissociation of BECLIN-1 from a BCL2-BECLIN-1 complex upon phosphorylation of BCL2 is an upstream signaling process for the induction of autophagy [12], we measured levels of phosphorylated BCL2 at serine 87 (p-BCL2Ser87), total BCL2, and BECLIN-1. Interestingly, both p-BCL2Ser87 and BCL2 levels were diminished, resulting in significant reduction in p-BCL2Ser87/BCL2 ratio in the absence of BECLIN-1 modulation in the EXE group (Figures 3a-3c). Moreover, ATG 7, a rate limiting enzyme involved in autophagy induction was repressed in response to EXE (Figures 3a and 3d). Next, we measured lysosome associated membrane protein-2 (LAMP-2) and CATHEPSIN L, both of which are linked to lysosomal fusion with autophagosomes and final degradation, respectively and found no significant changes after EXE (Figures 3a and 3e-3g).
Figure 3: Endurance exercise suppresses inductive processes of autophagy and maintains the same levels of lysosomal proteins in TA muscle. (a) Representative images of Western blot for p- BCL2Ser87, BCL2, BECLIN-1, ATG7, LAMP-2, and pro/active forms of CATHEPSIN L proteins in CON and EXE groups. (b-g) Quantitation of the intensity of each blot. Ponceau stained proteins were used as a loading control, and thus each target protein was normalized by the loading control. Error bar shows a mean ± SEM (n=7~9 per group). * p<0.05 vs. CON, ** p<0.01 vs. CON. CON: sedentary control group, EXE: endurance exercise group, TA: Tibialis Anterior, SEM: Standard Error of the Mean.
Endurance exercise promotes anabolic activities in TA muscle
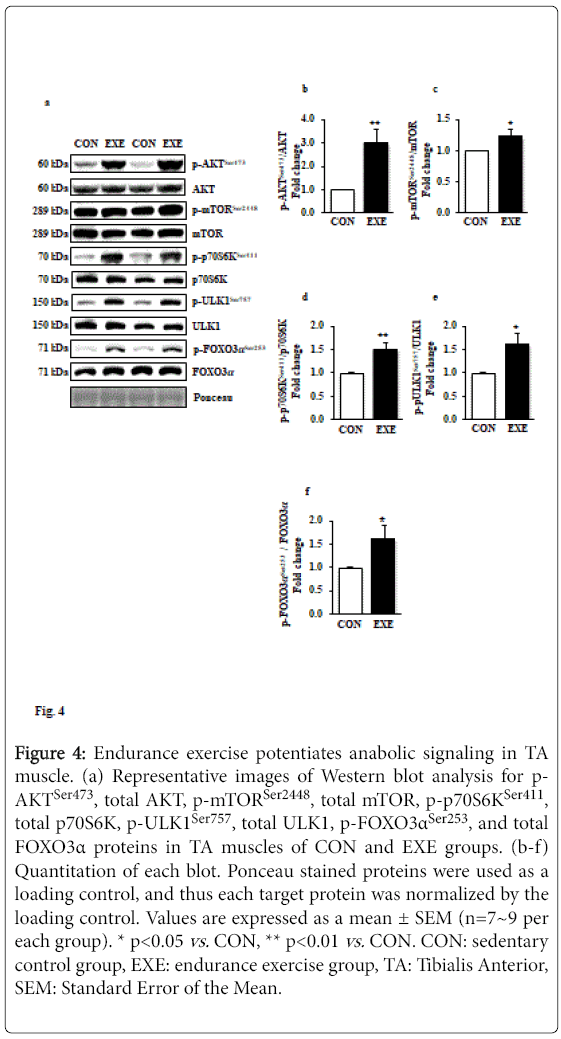
Elevated anabolic signaling pathways have been known to interfere in autophagy induction; for example, mTOR-induced phosphorylation of Unc-51 Like Kinase 1 (ULK1) prohibits class III PI3K activation and results in interrupting autophagy induction. To examine if EXE modulates anabolic signaling paradigms that may regulate activation or deactivation of autophagy, we assessed a series of canonical anabolic signaling molecules (e.g., AKT-mTOR-p70S6K axis). Our data revealed that phosphorylation levels of AKTSer47, mTORSer2448, and p70S6KSer411 (Figures 4a-d) were significantly elevated in response to EXE. Furthermore, phosphorylation levels of ULK1Ser757 by mTOR were increased (Figures 4a and 4e). We also observed that FOXO3, a key transcription factor associated with catabolism and autophagy, was hyperphosphorylated (inactivated) in response to exercise (Figures 4a and 4f).
Figure 4: Endurance exercise potentiates anabolic signaling in TA muscle. (a) Representative images of Western blot analysis for p- AKTSer473, total AKT, p-mTORSer2448, total mTOR, p-p70S6KSer411, total p70S6K, p-ULK1Ser757, total ULK1, p-FOXO3αSer253, and total FOXO3α proteins in TA muscles of CON and EXE groups. (b-f) Quantitation of each blot. Ponceau stained proteins were used as a loading control, and thus each target protein was normalized by the loading control. Values are expressed as a mean ± SEM (n=7~9 per each group). * p<0.05 vs. CON, ** p<0.01 vs. CON. CON: sedentary control group, EXE: endurance exercise group, TA: Tibialis Anterior, SEM: Standard Error of the Mean.
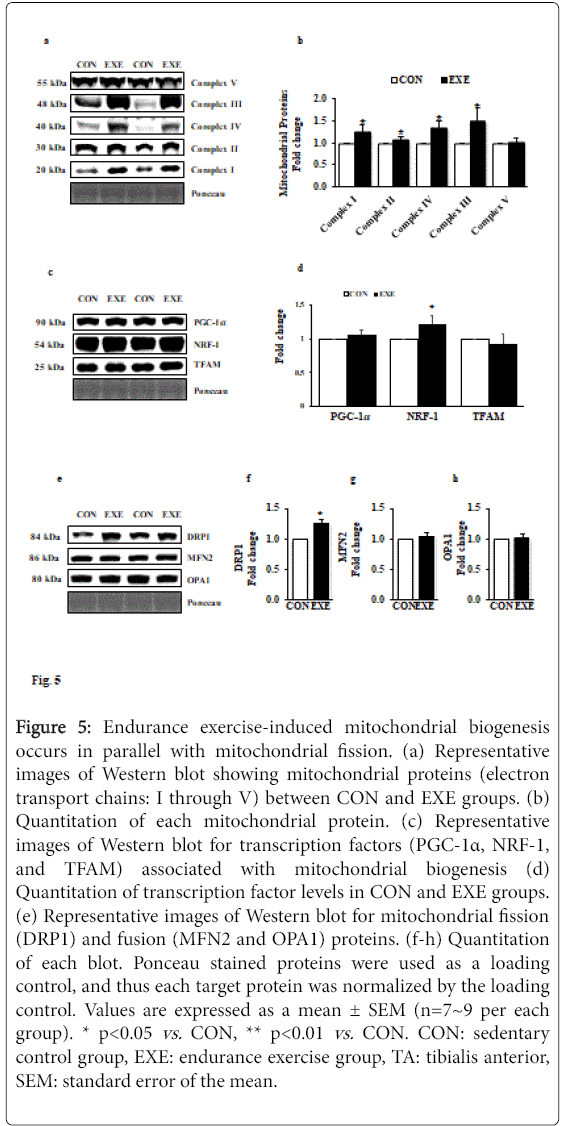
Endurance exercise induces mitochondrial biogenesis in TA muscle
To ascertain whether exercise-induced morphological modification (e.g., muscle mass reduction and fiber type shifts) is associated with mitochondrial biogenesis as well as mitochondrial turnover, we examined changes in five key mitochondrial proteins (complex I-V) and mitochondrial fusion and fission proteins (e.g., DRP1 for fission and MFN2 and OPA1 for fusion). Our data showed that all complex proteins (complex I through IV), except complex V, were significantly upregulated (Figures 5a and 5b). We next examined key mitochondrial biogenesis-related transcription factors. Intriguingly, neither peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) nor mitochondrial transcription factor A (TFAM) were changed; however, levels of nuclear respiratory factor-1 (NRF-1), another key transcription factor associated with mitochondrial biogenesis was upregulated (Figures 5c and 5d). Interestingly, while DRP1 levels were elevated (Figures 5e and 5f), MFN2 and OPA1 levels were not changed in response to EXE compared with CON group (Figures 5e, 5g and 5h).
Figure 5: Endurance exercise-induced mitochondrial biogenesis occurs in parallel with mitochondrial fission. (a) Representative images of Western blot showing mitochondrial proteins (electron transport chains: I through V) between CON and EXE groups. (b) Quantitation of each mitochondrial protein. (c) Representative images of Western blot for transcription factors (PGC-1α, NRF-1, and TFAM) associated with mitochondrial biogenesis (d) Quantitation of transcription factor levels in CON and EXE groups. (e) Representative images of Western blot for mitochondrial fission (DRP1) and fusion (MFN2 and OPA1) proteins. (f-h) Quantitation of each blot. Ponceau stained proteins were used as a loading control, and thus each target protein was normalized by the loading control. Values are expressed as a mean ± SEM (n=7~9 per each group). * p<0.05 vs. CON, ** p<0.01 vs. CON. CON: sedentary control group, EXE: endurance exercise group, TA: tibialis anterior, SEM: standard error of the mean.
Discussion
EXE is a potent inducer of autophagy in slow muscle tissues; however, it is not fully elucidated whether EXE induces autophagy in fast muscle tissues and if EXE-induced autophagy is associated with muscle fiber transformation. In the current study, we examined signaling nexus of autophagy pathways and biochemical as well as morphological adaptive responses in fast tibialis anterior (TA) muscles. In our study, three important findings arise. First, long-term EXE sustains a high level of autophagy in conjunction with emergence of aerobic phenotypes of TA muscle evidenced by reduced fiber crosssectional area, formation of slow muscle fibers, and enhanced mitochondrial biogenesis. Second, EXE suppresses an inductive process of autophagy, evidenced by diminished phosphorylation levels of both AMPK and BCL2, and ATG7 protein levels, despite elevated autophagy levels. Third, EXE potentiates anabolic signaling (AKTmTOR- p70S6K) pathways. Taken together, these findings suggest that EXE may serves as an important anabolic stimulator for transformation of muscle fiber types and mitochondrial biogenesis as well as modulator of autophagy flux.
Chronic EXE training has been used as a primary means to enhance muscular aerobic power which is acquired by transformation of fast (type IIx/b) to intermediate (type IIa) and to slow (type I) muscle fibers [27]. Similarly, our study showed that 6 weeks of EXE elicits an aerobic phenotype and reduced fiber cross-sectional area in TA muscle tissue. This morphological transformation is a beneficial adaptive response because reduced fiber sizes raises a capillary density even without an increase in number of capillaries and thus results in improved O2 delivery and enhanced aerobic capacity [6]. Although a clear mechanism of EXE-induced slimness of muscle fibers remain to be elucidated, autophagy has been reported to be associated with muscular transformation (e.g., oxidative muscle development) and metabolic reprogramming (oxidative phosphorylation) [12,17]. Supporting this, Yan et al. demonstrated that exercise-induced autophagy is an essential factor for developing aerobic capacity in skeletal muscle tissues, as perturbation of autophagy precludes muscle’s adaptive response to endurance exercise [17]. In this context, we also observed an increase in autophagy as evidenced by an elevated LC3-II level, an increase in LC3-II/I ratio, as well as BNIP3 after 6 weeks of EXE. Furthermore, we measured p62 to ensure if increased LC3-II levels reflect improved autophagy flux since it functions an adaptor protein in autophagosome formation and is degraded with autophagosomes by lysosomes [29]. While a reduced level of p62 has been accepted as reflection of improvement of autophagy flux [17,30], others have reported no changes in p62 levels, despite significant LC3- II accumulation. In our study, we also observed that p62 levels were unchanged in TA muscle in spite of LC3-II upregulation. More interestingly, recent studies have reported that p62 levels rather increases after endurance exercise in parallel with LC3-II elevation [17,27]. Currently, the reason for these discrepant findings (decrease vs. no change vs. increase in p62) remains unknown, and thus a genetic model study (e.g., p62 knockout) is necessary to resolve the conflict of whether modulation of p62 levels is an essential element for the assessment of EXE-induced autophagy.
Since autophagy is a lysosome-dependent catabolic process, proportional increases in lysosomal proteins LAMP2 and CATHEPSIN L in parallel with upregulated autophagosome formation [31]. Interestingly, however, several studies using EXE as an intervention have shown that LAMP2 [32] and CHAPTHEPSINS [33] were not changed. Consistent with these studies, our data also show that EXE does not modulate LAMP-2 and CATHEPSIN L levels, despite an increase in autophagy. Currently, it remains unknown whether a corresponding match between autophagosomes and lysosomal proteins for proper facilitation of autophagy flux is necessary. Thus, further studies need to elucidate this question.
Activation of AMPK induces autophagy, whereas a major anabolic effector, mTOR, opposes autophagy induction. More specifically, ULK1 plays a key role in transmitting autophagy induction signaling via activating class III PI3K; however, when ULK1 become phosphorylated at Ser757 by mTOR activation and its activity diminishes, and as a result, induction of autophagy is compromised [34]. In the current study, we found that EXE increases mTOR phosphorylation in parallel with AKT (an upstream molecule of mTOR) and p70S6K (a downstream molecule of mTOR) and that its elevation is associated with ULK1Ser757 phosphorylation. Collectively, our results suggest that long-term EXE may interrupt induction of autophagy by potentiating anabolic signaling in fast muscle tissues and possibly serve as a “off switch” to prevent chronic excessive autophagy that may occur in a situation where prolonged endurance exercise training (such as a 6-week long-term exercise protocol in our study) continues. In addition to reduced autophagy induction, we also found that the activity of a key catabolic mediator FOXO3α involved in expressing catabolic proteins [35] declines in EXE-trained TA muscle. FOXO3α is regulated by phosphorylation, and AKT is known to suppress its transcriptional activity by phosphorylating it [36]. Our data show that EXE-induced upregulation of FOXO3α phosphorylation concurs with enhanced AKT phosphorylation levels. This suggests that anabolic activation coincident with catabolic repression would be an important adaptive response of fast muscle tissues to prolonged endurance exercise.
Enhanced mitochondrial biogenesis in exercise-trained muscles is a key indicator of successful adaptation to endurance exercise [6,37,38]. Consistent with these studies, our results show EXE-induced mitochondrial biogenesis, as evidenced by upregulated mitochondrial proteins (e.g., electron transport chain: complex I-IV). Generally, mitochondrial biogenesis emerges with upregulation of key transcription factors such as PGC-1α, NRF1, and TFAM [39,40] ; interestingly, however, we did not observe upregulation of those proteins except for NRF1. Although it is not clear why upregulation of PGC-1α and TFAM are not observed in the current study, a recent study provides evidence that mitochondrial biogenesis can occur independent of PGC-1α [41]. It can be also presumed that duration of total training (e.g., acute vs. short-term vs. long-term) may affect the levels of those transcription factors; for example, if EXE-induced aerobic adaptation of muscle tissues reaches the pinnacle of mitochondrial biogenesis, transcription factors for mitochondrial biogenesis will return to basal levels to maintain cellular homeostasis. This may be one of possible explanations for the results of our study. Further studies are needed to examine the exact time periods of adaptive responses of mitochondrial biogenesis and homeostatic regulatory functions of key transcription factors of mitochondrial biogenesis.
Mitochondrial turnover (e.g., fusion and fission) is a critical element that is linked to mitochondrial biogenesis and elimination [42,43]. Although it is still unclear whether fission or fusion or both of them are linked to exercise-induced mitochondrial biogenesis, our findings, showing upregulation of mitochondrial fission protein DRP1, with fusion proteins OPA1 and MFN2 remaining unchanged, suggest that mitochondrial fission may be necessary for the biogenesis of mitochondria. In line with our data, recent studies also have revealed that endurance exercise enhances DRP1 expression without the alteration of MFN2 levels [30,42,44,45]. Nevertheless, given that mitochondria are highly dynamic organelles, and frequently undergoing fusion and fission processes in various stress conditions, our results (DRP1 upregulation) may not describe the entire adaptive outcomes of mitochondrial biogenesis in response to long-term EXE.
In conclusion, our findings support the general notion that longterm EXE is a key stimulator of transformation of muscular phenotypes: shifts from slow to fast muscle fibers, reduction in crosssectional area, and mitochondrial biogenesis in fast muscle tissues. Importantly, our results reveal that while long-term EXE retains increased autophagy (an increased in LC3-II and BNIP3 protein levels), autophagy induction is suppressed in parallel with enhanced anabolic signaling cascades (AKT-mTOR-p70S6K). This improved anabolic signaling cascade is associated with upregulation of mitochondrial biogenesis, which concurs with an increase in mitochondrial fission protein DRP1 in the absence of modulation of fusion proteins. Consequently, our findings provide an important insight into an adaptive mechanism of EXE-mediated autophagy in fast muscle tissues, supposing an anabolic signaling cascade as a possible feedback system necessary for guiding the suitable rate of autophagy in response to long-term EXE.
Acknowledgement
This project was supported by a grant from the University of West Florida through Office of Research and Sponsored Programs (R0062) and UWF Florida Research Fellowship to YL (CF6672).
References
- Akimoto T, Okuhira K,  Aizawa K, Wada S, Honda H, et al. (2013) Skeletal muscle adaptation in response to mechanical stress in p130cas-/- mice. Am J Physiol Cell Physiol 304: C541-547.
- Â Akimoto T, Ribar TJ, Williams RS, Yan Z (2004) Skeletal muscle adaptation in response to voluntary running in Ca2+/calmodulin-dependent protein kinase IV-deficient mice. Am J Physiol Cell Physiol 287: C1311-1319.
- Egan B, Zierath JR (2013) Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab 17: 162-184.
- Geng T, Â Li P, Okutsu M, Yin X, Â Kwek J, et al. (2010) PGC-1alpha plays a functional role in exercise-induced mitochondrial biogenesis and angiogenesis but not fiber-type transformation in mouse skeletal muscle. Â Am J Physiol Cell Physiol 298: C572-579.
- Rock KS, Hirshman MF, Brandauer J, Fujii N, Witters LA, et al. (2007) Skeletal muscle adaptation to exercise training: AMP-activated protein kinase mediates muscle fiber type shift. Diabetes 56: 2062-2069.
- Waters RE, Rotevatn S, Li P, Annex BH, Yan Z (2004) Voluntary running induces fiber type-specific angiogenesis in mouse skeletal muscle. Am J Physiol Cell Physiol 287: Â C1342-1348.
- Smiles WJ, Hawley JA, Camera DM (2016) Effects of skeletal muscle energy availability on protein turnover responses to exercise. J Exp Biol 219: 214-225.
- Ertunc M, Atalay A, Yildirim M, Onur R (2010) Exercise and suspension hypokinesia-induced alterations in mechanical properties of rat fast and slow-twitch skeletal muscles. Acta Physiol Hung 97: 316-325.
- Hwee DT, Kennedy AR, Hartman JJ, Ryans J, Durham N, et al.(2015) The small-molecule fast skeletal troponin activator, CK-2127107, improves exercise tolerance in a rat model of heart failure. J Pharmacol Exp Ther 353: 159-168.
- Shenkman BS, Belova SP, Lomonosova YN, Kostrominova TY, Nemirovskaya TL (2015) Calpain-dependent regulation of the skeletal muscle atrophy following unloading. Arch Biochem Biophys 584 : 36-41.
- Cunha TF, Moreira JB, Paixao NA, Campos JC, Monteiro AW, et al. (1985) Aerobic exercise training upregulates skeletal muscle calpain and ubiquitin-proteasome systems in healthy mice. J Appl Physiol 112: 1839-1846.
- He C, Bassik MC, Moresi V, Sun K, Wei Y (2012)Â Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 481: 511-515.
- Grumati P, Bonaldo P (2012) Â Autophagy in skeletal muscle homeostasis and in muscular dystrophies Cells 1: 325-345.
- Ryter SW, Cloonan SM, Choi AM (2013) Autophagy: A critical regulator of cellular metabolism and homeostasis. Mol Cells 36: 7-16.
- Dennemarker J, Lohmuller T, Muller S, Aguilar SV, Tobin DJ, et al. (2010) Impaired turnover of autophagolysosomes in cathepsin L deficiency. Biol Chem 391: 913-922.
- Huynh KK, Eskelinen EL, Scott CC, Malevanets A, Saftig P, et al. (2007) LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J 26: 313-324.
- Lira VA, Okutsu M, Zhang M, Greene NP, Laker RC, et al. (2013) Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance. FASEB J 27: 4184-4193.
- Nair U, Klionsky DJ (2011) Activation of autophagy is required for muscle homeostasis during physical exercise. Autophagy 7: 1405-1406.
- Masiero E, Sandri M (2010) Autophagy inhibition induces atrophy and myopathy in adult skeletal muscles. Autophagy 6: 307-309.
- Caberlotto L, Lauria M, Nguyen TP, Scotti M (2013) The central role of AMP-kinase and energy homeostasis impairment in Alzheimer's disease: a multifactor network analysis. PLoS One 8: e78919.
- Hao M, Zhu S, Hu L, Zhu H, Wu X, et al. (2017) Myocardial Ischemic Postconditioning promotes autophagy against ischemia reperfusion injury via the activation of the nNOS/AMPK/mTOR pathway. Int J Mol Sci 18: E614.
- Lantier L, Fentz J, Mounier R, Leclerc J, Treebak JT, et al. (2014) AMPK controls exercise endurance, mitochondrial oxidative capacity, and skeletal muscle integrity. FASEB J 28: 3211-3224.
- Lee Y, Kang EB, Kwon I, Cosio-Lima L, Cavnar P, et al. (2016) Cardiac kinetophagy coincides with activation of anabolic signaling. Med Sci Sports Exerc 48: 219-226.
- Sanchez AM, Csibi A, Raibon A, Cornille K, Gay S, et al. (2012) AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk1. J Cell Biochem 113: 695-710.
- Sun L, Zhang S, Yu C, Pan Z, Liu Y, et al. (2015) Hydrogen sulfide reduces serum triglyceride by activating liver autophagy via the AMPK-mTOR pathway. Am J Physiol Endocrinol Metab 309: E925-935.
- Sanchez AM, Candau RB, Csibi A, Pagano AF, Raibon A, et al. (2012) The role of AMP-activated protein kinase in the coordination of skeletal muscle turnover and energy homeostasis. Am J Physiol Cell Physiol 303: C475-485.
- Tam BT, Pei XM, Yu AP, Sin TK, Leung KK, et al. (2015) Autophagic adaptation is associated with exercise-induced fibre-type shifting in skeletal muscle. Acta Physiol (Oxf) 214: 221-236.
- Kammoun M, Cassar-Malek I, Meunier B, Picard B (2014) A simplified immunohistochemical classification of skeletal muscle fibres in mouse. Eur J Histochem 58: 2254.
- Mizushima N, Klionsky DJ (2007) Protein turnover via autophagy: Implications for metabolism. Annu Rev Nutr 27: 19-40.
- Southern WM, Nichenko AS, Shill DD, Spencer CC, Jenkins NT, et al.(2017) Skeletal muscle metabolic adaptations to endurance exercise training are attainable in mice with simvastatin treatment. PLoS One 12: e0172551.
- Medina DL, Paola S, Peluso I, Armani A, Stefani D, et al. (2015) Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 17: 288-299
- Kim YA, Kim YS, Oh SL, Kim HJ, Song W (2013) Autophagic response to exercise training in skeletal muscle with age. J Physiol Biochem 69: 697-705.
- Smuder AJ, Kavazis AN, Min K, Powers SK (2011) Exercise protects against doxorubicin-induced markers of autophagy signaling in skeletal muscle. J Appl Physiol 111: 1190-1198.
- Kim J, Kundu M, Viollet B, Guan KL (2011) AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13: 132-141.
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, et al. (2004) Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117: 399-412.
- Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, et al. (2004) Â The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell 14: 395-403.
- Geng T, Li P, Okutsu M, Yin X, Kwek J, et al. (2010) PGC-1alpha plays a functional role in exercise-induced mitochondrial biogenesis and angiogenesis but not fiber-type transformation in mouse skeletal muscle. Am J Physiol Cell Physiol 298: C572-579.
- Yan Z, Okutsu M, Akhtar YN, Lira VA (2011) Regulation of exercise-induced fiber type transformation, mitochondrial biogenesis, and angiogenesis in skeletal muscle. J Appl Physiol 110: 264-274.
- Kim JC, Park GD, Kim SH (2017) Inhibition of oxidative stress by antioxidant supplementation does not limit muscle mitochondrial biogenesis or endurance capacity in rats. J Nutr Sci Vitaminol 63: 277-283.
- Â Zoladz JA, Koziel A, Broniarek I, Woyda-Ploszczyca AM, Ogrodna K, et al. (2017) Effect of temperature on fatty acid metabolism in skeletal muscle mitochondria of untrained and endurance-trained rats. PLoS One 12: Â e0189456.
- Mansueto G, Armani A, Viscomi C, D'Orsi L, De Cegli R,et al.(2017) Transcription factor EB controls metabolic flexibility during exercise. Cell Metab 25: 182-196.
- Nichenko AS, Southern WM, Atuan M, Luan J, Peissig KB, et al. (2016) Mitochondrial maintenance via autophagy contributes to functional skeletal muscle regeneration and remodeling.  Am J Physiol Cell Physiol 311: C190-200.
- Youle RJ, van der Bliek AM (2012) Mitochondrial fission, fusion, and stress. Science 337: 1062-1065.
- Call JA, Wilson RJ, Laker RC, Zhang M, Kundu M, et al. (2017) Ulk1-mediated autophagy plays an essential role in mitochondrial remodeling and functional regeneration of skeletal muscle. Am J Physiol Cell Physiol 312: C724-C732.
- Pagano AF, Py G, Bernardi H, Candau RB, Sanchez AM (2014) Autophagy and protein turnover signaling in slow-twitch muscle during exercise. Med Sci Sports Exerc 46: 1314-1325.
Citation: Kwon I, Jang Y, Song W, M Cosio-Lima L, Lee Y (2018) Fast Twitch Skeletal Muscle Remodeling by Prolonged Endurance Exercise is Associated with Crosstalk between Anabolic and Catabolic Signaling Pathways in Mice . Biochem Physiol 7:228. DOI: 10.4172/2168-9652.1000228
Copyright: © 2018 Kwon I, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 4643
- [From(publication date): 0-2018 - Nov 28, 2024]
- Breakdown by view type
- HTML page views: 3920
- PDF downloads: 723