Research Article Open Access
Expression and Characterization of Capsid Proteins Derived from GII.17 and GII.7 Noroviruses
Xin Wan1#, Wenhui Wang1#, Yuqi Huo2#,Tong Ling1, Li Ding1, Jie Wu1, Shengli Meng1, Zejun Wang1* and Shuo Shen1*1Wuhan Institute of Biological Products, Wuhan P. R. China
2The Sixth People’s Hospital of Zhengzhou, Zhengzhou P. R. China
#These authors contributed equally
- *Corresponding Author:
- Zejun Wang
Wuhan Institute of Biological Products Co., Ltd. (WIBP)
No. 1 Golden Industrial Park Road
Zhengdian, Jiangxia Dist. Wuhan
430207, China
E-mail: wangzejun@sinopharm.com
Received date: July 26, 2016; Accepted date: September 09, 2016; Published date: September 20, 2016
Citation: Wan X, Wang W, Huo Y, Ling T, Ding L, et al. (2016) Expression and Characterization of Capsid Proteins Derived from GII.17 and GII.7 Noroviruses. J Emerg Infect Dis 1:113. doi:10.4172/2472-4998.1000113
Copyright: © 2016 Wan X, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Infectious Disease and Pathology
Abstract
Objective: Noroviruses (NoVs) are the leading cause of non-bacterial gastroenteritis worldwide and genogroup II, genotype 4 variants have been responsible for the majority of outbreaks reported. In winter 2014-15, the transient burst outbreaks caused by a new GII.17 variant that differed from previous strains was observed in several countries. In this study, major capsid proteins derived from GII.17 and GII.7 NoVs were expressed in sf9 cells and used to produce hyperimmune sera in rabbits for characterization of GII.17 and GII7 NoV Virus like particles (VLPs).
Methods: Baculovirus-insect cell expression system was used to assemble NoV VLPs. Salivary HBGA-viruslike particle (VLP) binding and binding blockade assays were performed.
Results: High expression levels were observed for both capsid proteins and expression led to successful assembly of VLPs. CsCl density gradient centrifugation indicated presence of two bands for both VLPs and SDS-PAGE analysis of these two bands showed contradictory ratio of the full-length and truncated capsid proteins. Both VLPs bound to salivary HBGAs derived from blood type A, B, AB and O individuals. In vitro HBGA-VLP binding blockade assay using hyperimmune sera against multiple genotypes, including current pandemic Sydney 2012 GII.4, GI.2, GII.7 and GII.17 VLPs demonstrated absence of blocking antibodies against the binding of GII.17 and GII.7 VLPs to salivary HBGAs.
Conclusion: The expression of GII.17 and GII.7 capsid proteins and VLP assembling allowed the antigenic comparison of different recombinant NoVs.
Keywords
NoVs; Histo-blood group antigens; VLPs; Crossblocking antibodies
Introduction
Since its recognition in 1972, Norovirus (NoV) has become one of the leading cause of non-bacterial gastroenteritis worldwide and leads to an average of 570 to 800 deaths annually [1-5]. NoVs can currently be divided at least into six genogroups and a tentative genogroup VII has been proposed [6]. Each genogroup is further divided into several genotypes and genogroup II, genotype 4 NoVs have been causing the majority of outbreaks worldwide [7-10]. Since 1995, periodically increasing activities of NoV have been associated with emergence of new GII.4 variants [11,12]. Available evidence indicates that the emergence of new endemic or pandemic variants might be due to escaping from protective herd immunity under immune-driven selection [11,13].
The absence of proper cell culture systems and small animal models has greatly hindered NoV-related pathophysiological studies and vaccine development. The discovery that expression of major capsid protein (VP1) using recombinant baculovirus expression system leads to assembly of virus-like particles has increased our understanding of NoV in several aspects, including serology, antibody cross-reactions among homologous and heterologous strains, virus structure and receptor/attachment factor identification [14-17].
The underlying mechanism of co-circulation of multiple genotypes with one particular genotype (GII.4) leading to pandemics is still not fully understood. It seems that such a discrepancy cannot be fully accounted for by the wide-spectrum tropism to varied histo-blood group antigens (HBGAs) as VLPs derived from other genotype also exhibit similar profile of HBGA binding [18,19]. The complexity of NoV infection is further deepened by the transient, spiked outbreaks caused by a new GII.17 variant that differed from previous strains [20-22]. Phylogenetic analysis indicated that the novel GII.17 strains clustered with strains isolated in 2012 in Kenya. The mechanism leading to sudden extreme prevalence of this new GII.17 strain is not clear. Other kinds of receptors or co-receptors might remain to be discovered and currently-used blocking of VLP-HBGAs should be further investigated to see if it represents the real neutralizing titers.
In this study, VP1 derived from a newly isolated GII.17 strain, as well as a GII.7 strain were expressed by using baculovirus expression system and used to produce hyperimmune sera in rabbits. In vitro salivary HBGA-VLP binding and binding blockade assays were performed to fully characterize the binding profiles of prepared VLPs and antibody blocking activities among multiple genotypes, including previously isolated strains in our lab [23].
Materials and Methods
Production of baculovirus recombinants
The full-length VP1 nucleotide sequences (Nt) derived from GII.17 and GII.7 (GenBank accession No. LC037415 and KJ196295, respectively) NoVs were codon-optimized, synthesized by Genscript company and inserted into transfer vector pBacPAK9 flanked by BamHI and NotI restricted enzyme sites at the 5’ and 3’ ends, respectively. The inserts were sequenced to be correct. Baculovirus recombinants were generated by following manufacturer’s instructions. In brief, purified, sterile vectors (2 μg) containing inserts were mixed with linearized baculovirus DNA (100 ng), followed by addition of Bacfectin (5 μl). Distilled water was added to the mixture to make a final volume of 100 μl, and after an incubation of 20 min at room temperature the mixture was added to six-well culture plate preseeded with Sf9 cells at a confluency of approximately 60-70%. The cells were cultured for 3-5 days before supernatant was harvested for infection of sf9 cells in T225 culture flasks. The medium was harvested 5-7 days post-infection and frozen at -80°C as viral stocks.
Purification of recombinant VLPs
The procedure for the purification of VLPs (GI.2, GII.3, GII.4, GII.7 and GII.17) was performed as previously reported [24]. Briefly, Sf9 cells cultured in suspension (serum-free medium) were infected with recombinant baculoviruses. The medium was generally harvested 5-7 days post-infection and then clarified by centrifugation at 10,000 xg for 30 min at 8°C. The VLPs in the supernatant were then pelleted by centrifugation at 141,000 xg for 3 h at 8°C in a SW 28 rotor. The VLPs in the pellet were resuspended in PBS (pH 7.2-7.4), mixed with equal volume of 1.6 g/ml CsCl and centrifuged at 288,000 xg for 24 h at 8°C in a SW 41 rotor. For isopycnic CsCl gradient centrifugation, visible bands were collected and ultracentrifuged at 141,000 xg for 3 h at 8°C in a SW 28 rotor. The purity and integrity of resuspended VLPs were analyzed by SDS-PAGE and verified by electromicroscopy (EM) after negative staining, respectively.
N-terminal sequencing of cleaved capsid proteins
For N-terminal amino acid sequence analysis, cleaved VP1was resolved by 10% separating gel and transferred to a PVDF membrane. The bands corresponding to cleaved fragments as confirmed in a Western blot analysis were excised. N-terminal microsequencing was performed on an N-terminal sequencer-PPSQ-33A.
Generation of hyperimmune sera in rabbits
Purified VLPs were quantified using BCA method and used to immunize Japanese big ear rabbits subcutaneously at a 2-week intervals for a total of four immunizations at 200 μg/dose mixed at 1:1 ratio with Freund’s complete adjuvant (first immunization) or Freund’s incomplete adjuvant (subsequent immunizations). Sera were collected one week after the fourth immunization and anti-VLP specific IgG titers were determined by using indirect enzyme-linked immunosorbent assay (ELISA).
ELISA
Cross-reactivity of rabbit anti-VLP antibodies against individual VLPs of different genotypes was determined by ELISA as described previously [25]. VLPs were coated onto 96-well microtiter plate wells at 2 μg/ml (100 μl/well). The end-point titers were calculated as reciprocal of the highest serum dilutions that gave an OD value greater than 0.1.
HBGA phenotyping of saliva samples
HBGAs in saliva samples collected from individuals with different blood types were characterized by using monoclonal antibodies against Lewis a, Lewis b, Lewis x, Lewis y, blood group A antigen, blood group B antigen, blood group H1 (O) antigen and blood group H2 antigen (Abcam, UK). In brief, saliva samples were boiled for 10 min to inactivate native antibodies. Saliva samples were then diluted in carbonate-bicarbonate buffer (pH 9.6) at 1:2,000 and coated onto 96- well microplates by incubating at 37°C overnight. The plates were then washed and blocked with PBS-T containing 1% BSA by incubating at 37°C for 1 h. The plates were washed five times with PBS-T and mouse anti-HBGA monoclonal antibodies diluted at 1:1,000 in PBS-T containing 1% BSA were added followed with an incubation period of 1 h at 37°C. After another five-times washing with PBS-T, Horse-radish peroxidase (HRP)-conjugated goat anti-mouse IgG or IgM antibody diluted at 1:2,000 was added. The plates were incubated at 37°C for 30 min. The color signal (450 nm) was detected by addition of HRP substrates tetramehthylbenzidine and urea peroxidase.
Saliva-VLP binding and binding blockade assay
Saliva-VLP binding and binding blockade assays were performed as described previously [24,26]. For both saliva-VLP binding and binding blockade assays, VLPs (GI.2, GII.3, GII.4, GII.7, GII.17) diluted in PBS-T containing 1% BSA at 0.25 μg/ml were used. For binding blockade assay, VLPs were pre-incubated with hyperimmune sera (rabbit anti-GI.2, GII.3, GII.4, GII.7 and GII.17 VLPs, respectively) for 30 min at 37°C before transferring to saliva precoated wells. The hyperimmune sera were diluted at 2-fold dilution starting from 1:800 to a maximum titer of 1:204,800.
Results
Expression of both VP1of GII.7 and GII.17 leads to assembly of VLPs
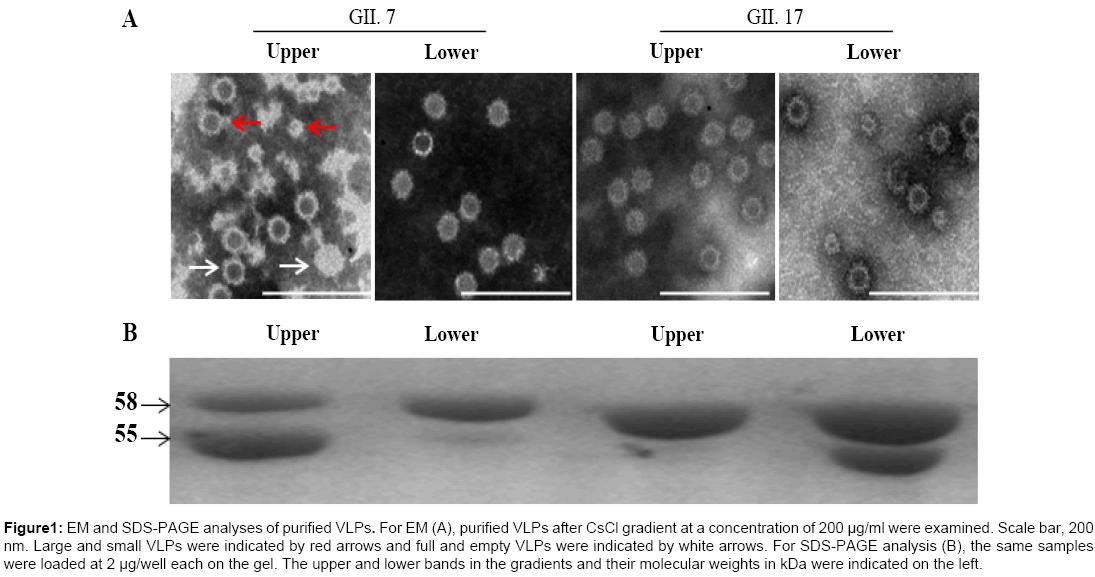
To increase protein expression level, both VP1 nucleotide sequences were optimized based on codon usage frequency in Spodoptera frugiperda (Sf9) cells followed by construction of transfer vectors. The final yields for GII.7 and GII.17 VLPs were approximately 33.1 mg/L and 128.2 mg/L, respectively, under suspension culture after purification (data not shown). EM analysis indicated that both VP1 successfully assembled into VLPs. As reported previously [25], both VP1 assembled VLPs exist in two forms, the larger VLPs similar to native, infectious virions in size and the smaller VLPs (Figure 1A).
Figure 1: EM and SDS-PAGE analyses of purified VLPs. For EM (A), purified VLPs after CsCl gradient at a concentration of 200 μg/ml were examined. Scale bar, 200 nm. Large and small VLPs were indicated by red arrows and full and empty VLPs were indicated by white arrows. For SDS-PAGE analysis (B), the same samples were loaded at 2 μg/well each on the gel. The upper and lower bands in the gradients and their molecular weights in kDa were indicated on the left.
Two bands exist for both VLPs after CsCl density gradient centrifugation
To obtain VLPs in high purity, CsCl density gradient centrifugation was employed. Two closely located bands were observed for both VLPs in the gradient. SDS-PAGE analysis of upper and lower bands for both VLPs indicated contradictory ratio of the 58 kDa full-length and the 55 kDa cleaved capsid proteins, with primarily the full-length capsid in the lower band for GII.7 VLPs and upper band for GII.17 VLPs. The cleaved capsid proteins were mainly located in the upper band for GII.7 VLPs and lower band for GII.17 VLPs (Figure 1B). For GII.7 VLPs, EM analysis indicated presence of some full-particles (particles with less electronic density) in the upper band compared with those in the lower band. For GII.17 VLPs, the full-particles were also observed in the upper band but at very low ratio to empty-particles (Figure 1A). Based on literature, the observed full-particles might be due to absorption of RNA to the surfaces of assembled VLPs.
Cleaved VP1 exhibits different N-terminal amino acid sequence
The truncated VP1 observed in this study was subject to N-terminal sequencing to determine the cleavage site. N-terminal sequencing results indicated that 38-aa was lost from the N-terminus of VP1 for GII.7 NoV (the first five aa of degraded protein was NH2-Thr-Pro-Val- Val-Gly) and inconclusive result was obtained for VP1 of GII.17 NoV, suggesting possible 34-38 amino acids lost from the N-terminus. The variation of different N-terminal sequences for cleaved VP1 has been reported, demonstrating non-specific cleavage and possible association with conformational changes of assembled VLPs [24,27,28].
Hyperimmune sera produced against GII.17 and GII.7 VLPs exhibit strong cross-reactivities with GII.4 VLPs in ELISA
The cross-reactivity of genotype-specific hyperimmune serum against all VLPs of 5 different genotypes was determined by ELISA (Table 1). The titers of rabbit anti-GII.7 VLPs were highest against GII.7 VLPs and 512-fold, 16-fold, 32-fold and 32-fold lower against GI.2, GII.4, GII.17 and GII.3 VLPs, respectively. The titers of rabbit anti-GII.17 VLPs were highest against GII.17 VLPs and 512-fold, 16- fold, 16-fold and 32–fold lower against GI.2, GII.4, GII.17 and GII.3 VLPs, respectively. Cross-reactivity between GII genotypes was at least 16-fold higher than that between genogroups I and II tested.
| VLP | ||||||
| Serum | Genetype | GI.2 | GII.3 | GII.4 | GII.7 | GII.17 |
| GI.2 | 1280 | 10 | 40 | 5 | 10 | |
| GII.3 | 1.25 | 1280 | 80 | 20 | 20 | |
| GII.4 | 0.625 | 10 | 1280 | 20 | 20 | |
| GII.7 | 10 | 160 | 320 | 5120 | 160 | |
| GII.17 | 5 | 80 | 160 | 160 | 2560 | |
Values in bold indicate titers against homologous VLPs. Titers represent the reciprocals of ELISA titers of sera tested against indicated VLPs and are in the unit of 104.
Table 1: Cross reactivity of hyperimmune sera produced against five different VLPs
Both VLPs bind to a wide spectrum of salivary HBGAs
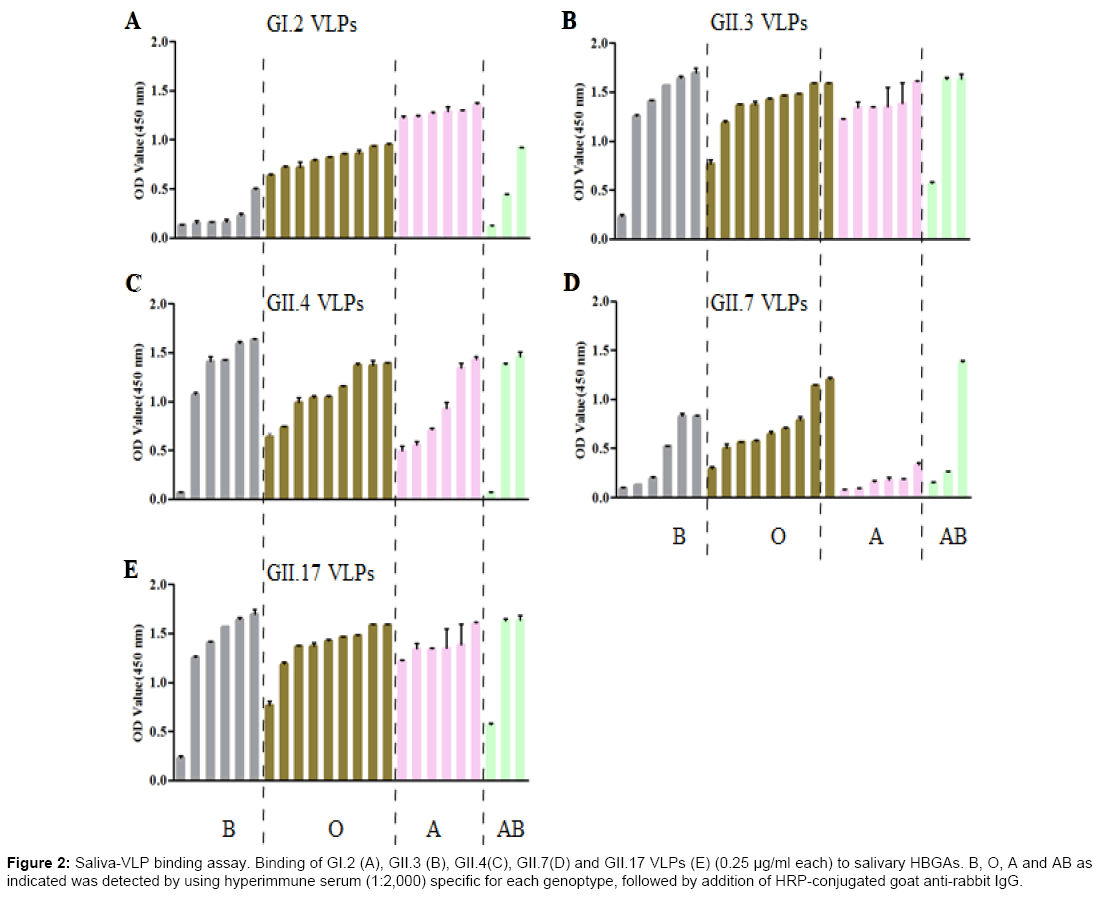
A total of 24 saliva samples collected from blood type A, B, AB and O individuals were used to characterize the binding profiles of prepared GII.7 and GII.17 VLPs, as well as previously reported GI.2, GII.3 and GII.4 VLPs. The HBGAs present in saliva samples were determined by using specific mouse anti-HBGAs monoclonal antibodies (mAb) (Table 2). The absence of H1 antigens in all saliva samples might be due to low sensitivity of mAb used or very low concentration of H1 antigens. As shown in Figure 2, GII.7 VLPs bound to all saliva samples with highest binding to blood type O, moderate binding to blood type B and lowest and similar binding signals to blood type A and AB salivary HBGAs. In comparison with GII.7 VLPs, GII.17 VLPs exhibited high and similar binding signals to all saliva samples. Using this larger panel of saliva samples, previously characterized GII.3 VLPs exhibited no binding to all saliva samples [25]. GI.2 VLPs showed highest binding signal to blood type A salivary HBGAs and GII.4 VLPs showed high and similar binding signals to blood type A, B, and O salivary HBGAs.
| HBGA* | ||||||||
| Saliva | Lea | Leb | A | H1 | H2 | Lex | Ley | B |
| B1 | - | + | - | - | - | - | + | +++ |
| B2 | - | + | - | - | - | - | + | +++ |
| B3 | + | + | - | - | - | - | + | +++ |
| B4 | + | - | - | - | - | - | + | +++ |
| B5 | + | +++ | - | - | - | - | + | +++ |
| B6 | + | - | - | - | - | - | - | ++ |
| O1 | - | +++ | - | - | - | - | + | - |
| O2 | + | +++ | - | - | - | - | + | + |
| O3 | + | +++ | - | - | - | - | + | + |
| O4 | + | +++ | - | - | - | - | + | - |
| O5 | + | +++ | - | - | - | - | + | - |
| O6 | - | ++ | - | - | + | - | - | - |
| O7 | + | +++ | - | - | - | - | + | |
| O8 | ++ | +++ | - | - | + | - | + | - |
| O9 | + | +++ | - | - | - | - | + | - |
| A1 | ++ | +++ | +++ | - | - | - | + | - |
| A2 | ++ | +++ | +++ | - | - | - | + | - |
| A3 | + | +++ | ++ | - | - | - | + | - |
| A4 | + | +++ | ++ | - | - | - | + | - |
| A5 | - | + | ++ | - | - | - | + | - |
| A6 | + | ++ | ++ | - | - | - | + | + |
| AB1 | ++ | +++ | - | - | - | - | + | - |
| AB2 | + | + | - | - | - | - | + | +++ |
| AB3 | + | + | - | - | - | - | - | ++ |
*:+, 0.1 < OD<0.5; ++, 0.5 < OD < 1.0; +++, OD > 1.0; -, OD < 0.1. A: blood type A; B: blood type B; O: blood type O; AB: blood type AB.
Table 2: HBGA phenotyping of saliva samples
Figure 2: Saliva-VLP binding assay. Binding of GI.2 (A), GII.3 (B), GII.4(C), GII.7(D) and GII.17 VLPs (E) (0.25 μg/ml each) to salivary HBGAs. B, O, A and AB as indicated was detected by using hyperimmune serum (1:2,000) specific for each genoptype, followed by addition of HRP-conjugated goat anti-rabbit IgG.
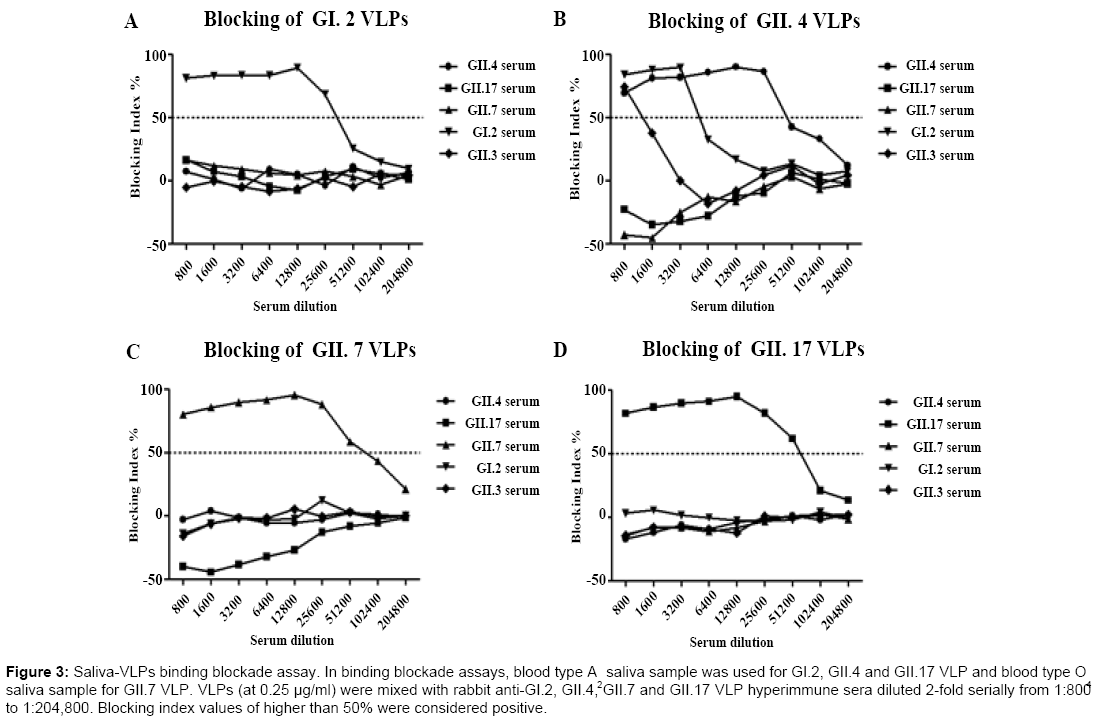
Absence of cross-blocking activity among different genotype antibody
In vitro binding blockade assay was performed to determine crossblocking activities of hyperimmune sera produced against different genotypes. Blood type A2 saliva sample was used for GI.2, GII.4 and GII.17 VLP binding blockade assay and blood type O4 saliva sample for GII.7 VLP binding blockade assay. The binding blockade assay was optimized for each VLP. As shown in Figure 3, binding of GI.2, GII.7 and GII.17 VLPs to salivary HBGAs could only be blocked by corresponding genotype-specific hyperimmune serum samples while binding of GII.4 VLPs to salivary HBGAs could be blocked by rabbit anti-GI.2, GII.4 and GII.3 hyperimmune sera at a dilution of 1:800, which was in consistency with our previous study [25]. The blocking activities against the binding of GII.4 VLPs to salivary HBGAs for hyperimmne sera against GI.2 and GII.3 NoV VLPs rapidly decreased when dilutions were greater than 800-fold and 3200-fold, respectively.
Figure 3: Saliva-VLPs binding blockade assay. In binding blockade assays, blood type A saliva sample was used for GI.2, GII.4 and GII.17 VLP and blood type O saliva sample for GII.7 VLP. VLPs (at 0.25 μg/ml) were mixed with rabbit anti-GI.2, GII.4, GII.7 and GII.17 VLP hyperimmune sera diluted 2-fold serially from 1:800 to 1:204,800. Blocking index values of higher than 50% were considered positive.
Discussion
A better understanding of epidemiological, biological and immunological profiles of dominating NoV strains is important for NoV vaccine development. The currently tested NoV vaccine candidates have been targeting the dominating GII.4 strains and prototype Norwalk virus, a GI.1 strain [29]. Increased cases caused by a GII.17 NoV strain during 2014/15 winter season in China and Japan was observed and in some areas this newly emerged GII.17 strain has replaced GII.4 Sydney 2012 strain as the dominating strain [21]. To fully characterize the binding profiles of this newly emerged GII.17 strain, its VP1 was expressed, purified and used to produce hyperimmune serum. At the same time, VP1 derived from a GII.7 strain was also expressed and corresponding hyperimmune serum produced for comparison in this study.
Purification of NoV VLPs using CsCl density gradient centrifugation generally gives two visible bands in proximity, a upper band composed of the full-length and truncated VP1 and a lower band composed of the full-length VP1. EM examination of both bands reveals regular VLPs in the upper band and irregular smear VLPs with blurred shape in the lower band (data not shown). For GII.7 and GII.17 NoV VLPs, two bands were also observed in CsCl density gradient centrifugation. SDS-PAGE analysis indicated contradictory locations of the full-length and truncated VP1. EM analysis indicated presence of similar VLPs in size and morphology within both bands but minor differences existed. If full-particles observed under EM were indeed due to absorption of RNA as reported [30], the higher ratio of the full-particles to the empty particles and higher percentage of truncated VP1 in the upper band for GII.7 VLPs indicated that absorption of nucleic acids could not prevent cleavage or degradation of the N-terminal amino acids, or adsorption of RNA happened after cleavage or degradation. The contradictory locations of the full-length and truncated VP1 for GII.17 VLPs seems cannot be explained by the presence of full- or empty-particles. It should be noted that bands composed of the full-length VP1 for both GII.7 and GII.17 VLPs exhibited dark background under EM, indicating presence of carbohydrates or lipids. The presence of salts can be ruled out as bands harvested from CsCl gradients were further ultracentrifuged by first diluting with 0.01 M PBS. Based on above observation, it is assumed that the observed unidentified substances might be able to prevent cleavage or degradation of assembled VLPs. In fact, it has been reported that HBGA-like substance-expressing bacteria can increase the stability of NoVs under heat stress [31]. It should be emphasized that more studies should be performed using the fecal samples of patients to see whether the different forms of NoV particles exist after infection of human.
Cleavage or degradation of the N-terminal amino acids of VP1 has been observed for both GI and GII NoV strains and such cleavage or degradation can be inhibited to a certain degree with serum-containing culture medium, indicating that serum contains enzyme inhibitors. Combining available information relating to varied N-terminal amino acid sequences reported and observed in this study for truncated VP1, it is assumed that the N-terminal amino acids are subject to non-specific degradation. Such degradation has been observed in partially purified wild type NoVs. In our recent studies, NoV-positive fecal samples (GII.3 and GII.4) were first clarified to remove insoluble material and then ultracentrifuged to pellet virus. Western blot analysis of pelleted virus indicated presence of two bands similar to those observed for prepared VLPs. EM analysis indicated presence of empty-particles with the morphology of NoV VLPs. Based on X-ray crystallography of NoV VLPs, the N-terminus was buried inside assembled VLPs [15,32]. The susceptibility of the N-terminal amino acids to enzymes indicates assembled VLPs are possibly undergoing conformational change.
Compared with GII.4 NoV, VLPs derived from GII.17 NoV exhibited similar and high binding signals to all saliva samples tested and such wide-spectrum binding activity might be associated with its emergence and rapid transmission [22]. VLPs derived from GII.7 NoV showed similar binding patterns with that of GII.4 NoV. Compared with the pandemic GII.4 strains, the low prevalence of GII.7 NoV, as well as restricted circulation area of GII.17 NoVs hint that other factors are at play for efficient transmission of these strains. Data from our ongoing epidemiological study of NoVs in Jingzhou, Hubei province and Zhengzhou, Henan province indicate that Sydney 2012 GII.4 variant is still the dominating strain, followed by GII.3 strains. Current HBGAs binding assay uses saliva samples, which might not reflect the actual intestinal environment, especially with the finding that HBGAlike substance present on bacteria can bind NoV VLPs and promote NoV infection of B cells [33]. Therefore, the low binding activities of certain genotype derived VLPs to saliva samples cannot be simply used to explain their restricted circulation and low prevalence. One handy example is that VLPs derived from GII.3 strains showed limited and weak binding activities to most saliva samples, which is contradictory to its high prevalence. Comparative evolutionary study has suggested that RdRp acquired by intergenic recombination might be a major driving force for the increased diversity and fitness of GII.3 NoVs [34].
The effectiveness of in vitro VLP-HBGAs binding blockade assay as surrogate neutralization assay has been recently put to test following with several phase I clinical trials. Good correlations have been observed between high HBGA blocking titers and protection of clinical gastroenteritis [35,36]. However, some participants without measurable anti-NoV VLPs antibodies pre-challenge were not infected after challenge with wild type virus, indicating other factors can influence the susceptibility status of certain individuals. In this study, cross-genotype blocking activities were not observed except for onesided blocking activity for rabbit anti-GI.2 and GII.3 hyperimmune serum as has been reported in our previous study [25]. Serological study using acute phase serum collected from a child infected with GII.4 Sydney 2012-like strain showed that the serum sample blocked the binding of GII.17 NoV VLPs to blood type A, B, AB and O saliva samples, but not GI.2, GII.4 and GII.7 NoV VLPs. The absence of crossblocking activities among different genotypes, as well as recent sudden burst outbreaks caused by GII.17 NoV strain indicated that NoV vaccines composed of multiple genotype-specific VLPs are required.
References
- Kapikian AZ, Wyatt RG, Dolin R, ThornhillTS, Kalica AR, et al. (1972) Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J Virol10: 1075-1081.
- Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinje J, et al. (2008) Systematic literature review of role of noroviruses in sporadic gastroenteritis.Emerg Infect Dis14: 1224-1231.
- Glass RI, ParasharUD, Estes MK (2009) Norovirus gastroenteritis. N Engl J Med361: 1776-1785.
- Hall AJ, Lopman BA, Payne DC, Patel MM, Gastanaduy PA, et al. (2013) Norovirus disease in the United States. Emerg Infect Dis19: 1198-1205.
- Mead PS, Slutsker L, Dietz V, McCaig LF, BreseeJS, et al. (1999) Food-related illness and death in the United States. Emerg Infect Dis5: 607-625.
- Vinje J (2015) Advances in laboratory methods for detection and typing of norovirus.J ClinMicrobiol53: 373-381.
- Zheng DP, Widdowson MA, Glass RI, Vinje J (2010) Molecular epidemiology of genogroup II-genotype 4 noroviruses in the United States between 1994 and 2006. J ClinMicrobiol48: 168-177.
- Thongprachum A, Khamrin P, Maneekarn N, Hayakawa S, Ushijima H (2016) Epidemiology of gastroenteritis viruses in Japan: Prevalence, seasonality, and outbreak. J Med Virol88: 551-570.
- Mans J, Murray TY, Nadan S, Netshikweta R, Page NA, et al. (2016) Norovirus diversity in children with gastroenteritis in South Africa from 2009 to 2013: GII.4 variants and recombinant strains predominate.Epidemiol Infect144: 907-916.
- Cho HG, Park PH, Lee SG, Kim JE, Kim KA, et al. (2015) Emergence of Norovirus GII.4 variants in acute gastroenteritis outbreaks in South Korea between 2006 and 2013. J ClinVirol72: 11-15.
- Lindesmith LC, Donaldson EF, Lobue AD, Cannon JL, Zheng DP, et al. (2008) Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med5: e31.
- Eden JS, Hewitt J, Lim KL, Boni MF, Merif J, et al. (2014) The emergence and evolution of the novel epidemic norovirus GII.4 variant Sydney 2012. Virology450: 106-113.
- Bull RA, White PA (2011) Mechanisms of GII.4 norovirus evolution.Trends Microbiol19: 233-240.
- Gray JJ, Jiang X, Morgan-Capner P, Desselberger U, Estes MK (1993) Prevalence of antibodies to Norwalk virus in England: detection by enzyme-linked immunosorbent assay using baculovirus-expressed Norwalk virus capsid antigen. J ClinMicrobiol31: 1022-1025.
- Prasad BV, Rothnagel R, Jiang X, Estes MK (1994) Three-dimensional structure of baculovirus-expressed Norwalk virus capsids. J Virol68: 5117-5125.
- Marionneau S, Ruvoen N, Le Moullac-Vaidye B, Clement M, Cailleau-Thomas A, et al. (2002) Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology122: 1967-1977.
- TreanorJJ, Jiang X, Madore HP, Estes MK (1993) Subclass-specific serum antibody responses to recombinant Norwalk virus capsid antigen (rNV) in adults infected with Norwalk, Snow Mountain, or Hawaii virus. J ClinMicrobiol31: 1630-1634.
- Huang P, Farkas T, Zhong W, Tan M, Thornton S, et al. (2005) Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns.J Virol79: 6714-6722.
- Huang P, Farkas T, Marionneau S, Zhong W, Ruvoen-Clouet N, et al. (2003) Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J Infect Dis188: 19-31.
- Matsushima Y, Ishikawa M, Shimizu T, Komane A, Kasuo S, et al. (2015) Genetic analyses of GII.17 norovirus strains in diarrheal disease outbreaks from December 2014 to March 2015 in Japan reveal a novel polymerase sequence and amino acid substitutions in the capsid region. Euro Surveill20.
- De Graaf M, van Beek J, Vennema H, Podkolzin AT, Hewitt J, et al. (2015) Emergence of a novel GII.17 norovirus - End of the GII.4 era?Euro Surveill20.
- Lu J, Sun L, Fang L, Yang F, Mo Y, et al. (2015) Gastroenteritis Outbreaks Caused by Norovirus GII.17, Guangdong Province, China, 2014-2015. Emerg Infect Dis21: 1240-1242.
- Huo Y, Cai A, Yang H, Zhou M, Yan J, et al. (2014) Complete nucleotide sequence of a norovirus GII.4 genotype: evidence for the spread of the newly emerged pandemic Sydney 2012 strain to China. Virus Genes48: 356-360.
- Huo Y, Wan X, Wang Z, Meng S, Shen S (2015) Production of NorovirusVLPs to size homogeneity. Virus Res204: 1-5.
- Huo Y, Wan X, Ling T, Shen S (2016) Biological and immunological characterization of norovirus major capsid proteins from three different genotypes. MicrobPathog90: 78-83.
- Huo Y, Wan X, Ling T, Wu J, Wang Z, et al. (2015) Prevailing Sydney like Norovirus GII.4 VLPs induce systemic and mucosal immune responses in mice. Mol Immunol68: 367-372.
- Koho T, Huhti L, Blazevic V, Nurminen K, Butcher SJ, et al. (2012) Production and characterization of virus-like particles and the P domain protein of GII.4 norovirus. J Virol Methods179: 1-7.
- Belliot G, Noel JS, Li JF, Seto Y, Humphrey CD, et al. (2001) Characterization of capsid genes, expressed in the baculovirus system, of three new genetically distinct strains of "Norwalk-like viruses". J ClinMicrobiol39: 4288-4295.
- TreanorJJ, AtmarRL, Frey SE, Gormley R, Chen WH, et al. (2014) A novel intramuscular bivalent norovirus virus-like particle vaccine candidate--reactogenicity, safety, and immunogenicity in a phase 1 trial in healthy adults. J Infect Dis210: 1763-1771.
- Kumar S, Ochoa W, Kobayashi S, Reddy VS (2007) Presence of a surface-exposed loop facilitates trypsinization of particles of Sinsiro virus, a genogroup II.3 norovirus. J Virol81: 1119-1128.
- Li D, Breiman A, le Pendu J, Uyttendaele M (2015) Binding to histo-blood group antigen-expressing bacteria protects human norovirus from acute heat stress. Front Microbiol6: 659.
- Prasad BV, Hardy ME, Dokland T, Bella J, Rossmann MG, et al. (1999) X-ray crystallographic structure of the Norwalk virus capsid. Science286: 287-290.
- Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, et al. (2014) Enteric bacteria promote human and mouse norovirus infection of B cells. Science346: 755-759.
- Mahar JE, Bok K, Green KY, Kirkwood CD (2013) Theimportance of intergenic recombination in norovirus GII.3 evolution. J Virol87: 3687-3698.
- Reeck A, Kavanagh O, Estes MK, Opekun AR, Gilger MA, et al. (2010) Serological correlate of protection against norovirus-induced gastroenteritis.J Infect Dis202: 1212-1218.
- AtmarRL, Bernstein DI, Lyon GM, TreanorJJ, Al-Ibrahim MS, et al. (2015) Serological Correlates of Protection against a GII.4 Norovirus.Clin Vaccine Immunol22: 923-929.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 11923
- [From(publication date):
September-2016 - Mar 29, 2025] - Breakdown by view type
- HTML page views : 10979
- PDF downloads : 944