Research Article Open Access
Evaluation of Phenotyping and Genotyping Characteristic of Shigella sonnei after Biofield Treatment
Mahendra Kumar Trivedi1, Shrikant Patil1, Harish Shettigar1, Khemraj Bairwa2 and Snehasis Jana2*1Trivedi Global Inc., 10624 S Eastern Avenue Suite A-969, Henderson, NV 89052, USA
2Trivedi Science Research Laboratory Pvt. Ltd., Hall-A, Chinar Mega Mall, Chinar Fortune City, Hoshangabad Rd., Bhopal- 462026, Madhya Pradesh, India
- Corresponding Author:
- Snehasis Jana
Trivedi Science Research Laboratory Pvt. Ltd.
Hall-A, Chinar Mega Mall, Chinar Fortune City, Hoshangabad Rd.
Bhopal- 462026, Madhya Pradesh, India
Tel: +91-755-6660006
E-mail: publication@trivedisrl.com
Received date: August 05, 2015; Accepted date: August 25, 2015; Published date: September 01, 2015
Citation: Trivedi MK, Patil S, Shettigar H, Bairwa K, Jana S (2015) Evaluation of Phenotyping and Genotyping Characteristic of Shigella sonnei after Biofield Treatment. J Biotechnol Biomater 5:196. doi:10.4172/2155-952X.1000196
Copyright: © 2015 Trivedi MK, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
Shigella sonnei (S. sonnei) is a non-motile, rod shape, clinically significant, Gram-negative bacterium. It is commonly associated with dysentery (shigellosis). Recently, resistance to third and fourth generation cephalosporins and fluoroquinolones has been reported in S. sonnei. In the present study, we assessed the effect of biofield treatment on phenotyping and genotyping characteristic of S. sonnei (ATCC 9290). The lyophilized samples of S. sonnei were divided in three groups (G): G-I (control, revived), G-II (treatment, revived), and G-III (treatment, lyophilized). All these groups (control and biofield treated) were analyzed against antimicrobial susceptibility, biochemical reactions, and biotype number. The 16S rDNA sequencing was carried out to establish the phylogenetic relationship of S. sonnei with different bacterial species. The treated cells of S. sonnei exhibited an alteration of 3.33%, 10%, and 23.33% of total 30 tested antimicrobials in susceptibility assay for G-II on day 5 and 10 and G-III on day 10, respectively as compared to control. The treated cells of S. sonnei showed a significant change of about 12.12%, 12.12%, and 57.58% biochemical reactions out of 33 tests in treated groups of G-II on day 5 and 10 and G-III on day 10, respectively. The biotype number was also changed in treated samples of S. sonnei. Based on nucleotide homology sequences and phylogenetic analysis, the nearest homolog species of S. sonnei (GenBank Accession Number: EU009190) was identified as Shigella flexneri (EF643608). These results revealed that biofield treatment can prevent the absolute resistance in microbe against the existing antimicrobials.
Keywords
Antimicrobial susceptibility; Biofield treatment; 16S rDNA gene sequencing; Shigella sonnei
Abbreviations
MIC: Minimum Inhibitory Concentration; ATCC: American Type Culture Collection; NBPC30: Negative Breakpoint Combo 30; NCBI: National Center for Biotechnology Information; WHO: World Health Organization; 16S rDNA: 16Svedberg Unit Ribosomal Deoxyribonucleic Acid; BLAST: Basic Local Alignment Search Tool; Outs: Operational Taxonomic Units
Introduction
Development of antimicrobial resistance in several microbes like bacteria, viruses, fungi, or in parasites has been reported globally in the recent few decades. Frequent and improper use of antimicrobial further accelerated the incidence of microbial resistance [1]. Shigella sonnei (S. sonnei) is a rod shape, non-motile, facultative anaerobic Gram-negative and lactose-fermenting bacterium. S. sonnei is associated with gastrointestinal tract (GIT) infection disease shigellosis in both developed and developing countries, where the sanitation is insufficient [2,3]. S. sonnei is usually transmitted by fecal-oral route, direct interpersonal contact, contaminated food, water, or uncooked food. Shigella infection is the third most common gastroenteritis after Salmonella and Campylobacter infection in the USA. Recently, S. sonnei has become the most prevalent species in the developed world. It is estimated to cause 80–165 million cases of disease and 600,000 deaths annually, worldwide [4]. The S. sonnei has been acquired resistant to commonly used antimicrobials like streptomycin, tetracycline, sulfonamide, trimethoprim, and ampicillin. Emergence of extendedspectrum β-lactamases (ESBLs) in S. sonnei was also detected in Korea [2,5]. Therefore the multidrug therapy required to treat the infection cause by resistant strain of microbes. However, multiple drug therapy shows serious toxicity and associated adverse effects like neurotoxicity and nephrotoxicity [6]. Due to associated side effects and failure of drug therapy, an alternate treatment approach is required. Recently, an alternate treatment known as biofield energy is reported that inhibits the growth of bacterial cultures [7]. Biofield is an electromagnetic field that permeates and surrounds living organisms and referred as biologically produced electromagnetic and subtle energy field that provides regulatory and communication functions within the human organism [8]. Various internal physiological processes such as blood flow, brain and heart function etc. generates biofield. Researchers have attempted different biologic studies and effects of biofield on various biomolecules such as proteins, antibiotics [9], conformational change in DNA [10] etc. Thus, human has the ability to harness the energy from environment or universe and can transmit into any living or nonliving object(s) around the Globe. The objects always receive the energy and responding into useful way that is called biofield energy and the process is known as biofield treatment [11]. Mr. Mahendra Trivedi’s biofield treatment (The Trivedi Effect®) has renowned to alter the various physicochemical characteristics of metals and ceramics [11-17]. Quality and quantity of several agriculture products have been improved by several folds in the biofield treated plants [18-20] and growth and adaptation of the plant were also enhanced with the help of biofield treatment [21,22]. In addition, the biofield treatment has considerably altered the phenotype and biotype of the microbe and subsequently, the susceptibility to antimicrobials was also changed [23-25].
Based on the knowledge of existing literatures and considering the clinical significance of S. sonnei, we evaluated to see the impact of biofield treatment on antimicrobial susceptibility, biochemical reactions pattern, biotype number, and 16S rDNA gene sequencing of the microbe.
Materials and Methods
Two lyophilized vials of S. sonnei [American Type Culture Collection (ATCC) 9290] were purchased from MicroBioLogics, Inc., USA. The microbial sample vials were stored as per the suggested storage conditions till further use. The antimicrobial susceptibility study, biochemical reactions pattern, and biotype number were evaluated by MicroScan Walk-Away® (Dade Behring Inc., West Sacramento, CA) through Negative Breakpoint Combo 30 (NBPC30) panel. The 16S rDNA sequencing was performed using Ultrapure Genomic DNA Prep Kit (Cat KT 83, Bangalore Genei, India).
Biofield treatment
The lyophilized strain of S. sonnei were divided into three groups (G) like G-I (control), G-II (treatment, revived), and G-III (treatment, lyophilized). G-I consider as control. No treatment was given. The treatment groups (II and III) were in sealed pack and handed over to Mr. Trivedi for biofield treatment under laboratory condition. Mr. Trivedi provided the treatment through his energy transmission process to the treated groups without touching the samples. Subsequently, group G-I and G-II were assessed on day 5 and 10; and G-III was assessed on day 10. After that, all groups were evaluated for an antimicrobial susceptibility, biochemical reactions pattern, and biotype number [25]. The 16S rDNA gene sequencing of S. sonnei was also carried out.
Investigation of antimicrobial susceptibility of S. sonnei
The antimicrobial susceptibility of S. sonnei was evaluated with the help of automated instrument, MicroScan Walk-Away® using Negative Breakpoint Combo 30 (NBPC30) panel as per the manufactures instructions [26]. The minimum inhibitory concentration (MIC) and a qualitative susceptibility like resistant (R), intermediate (I), and susceptible (S) were determined by analyzing the lowest antimicrobial concentration showing microbial growth inhibition [25]. The antimicrobial sensitivity study was carried out using following 30 antimicrobials such as amikacin, amoxicillin/K-clavulanate acid, ampicillin/sulbactam, ampicillin, aztreonam, cefazolin, cefepime, cefotaxime, cefotetan, cefoxitin, ceftazidime, ceftriaxone, cefuroxime, cephalothin, chloramphenicol, ciprofloxacin, gatifloxacin , gentamicin, imipenem, levofloxacin, meropenem, moxifloxacin, nitrofurantoin, norfloxacin, piperacillin, tazobactam, tetracycline, ticarcillin, tobramycin, and trimethoprim/sulfamethoxazole. All these antimicrobials were procured from Sigma-Aldrich.
Biochemical studies
The biochemical reactions of S. sonnei were carried out using MicroScan Walk-Away® system where, interpretation of biochemical reactions for microbial identification of Gram-negative organisms resulted in high accuracy [27,28]. The biochemical reactions patterns of control and treated samples of S. sonnei were performed using the following 33 biochemicals such as acetamide, adonitol, arabinose, arginine, cetrimide, cephalothin, citrate, colistin, esculin hydrolysis, nitrofurantoin, glucose, hydrogen sulfide, indole, inositol, kanamycin, lysine, malonate, melibiose, nitrate, oxidation-fermentation, galactosidase, ornithine, oxidase, penicillin, raffinose, rhamnose, sorbitol, sucrose, tartrate, tryptophan deaminase, tobramycin, urea, and Voges-Proskauer. All these biochemicals were procured from Sigma-Aldrich.
Biotype number
The biotype number of S. sonnei was found out utilizing the MicroScan Walk-Away® processed panel data report, using biochemical reactions data [26].
16S rDNA gene sequencing
Genomic DNA was prepared from biofield treated S. sonnei cells using genomic purification Kit, as per the manufacturer’s instructions. Subsequently, the 16S rDNA gene (~1.5 kb) was amplified using universal forward primer 5'-AGAGTTTGATCCTGGC-3' and universal reverse primer 5'-GGTTACCTTGTTACGACTT-3'. Subsequently, the amplified products were resolved by gel electrophoresis on 1.0% agarose gel, stained with ethidium bromide, and then visualized under UV light in a gel documentation unit (BioRad Laboratories, USA). The PCR amplified fragment was purified from the agarose gel utilizing a DNA Gel Extraction Kit. The amplified product was sequenced on commercial basis from Bangalore Genei, India. The received 16S rDNA sequences data were aligned and compared with the sequences stored in Gene Bank database of National Center for Biotechnology Information (NCBI) using the algorithm BLASTn program. Finally, the multiple sequence alignment/phylogenetic tree was constructed with help of MEGA 3.1 software utilizing neighbor joining method [29,30].
Results
Antimicrobial susceptibility assay
The antimicrobial sensitivity data were reported in Table 1 and 2. The result showed that the biofield treated S. sonnei exhibited a significant alteration in susceptibility assay of about 3.33% (G-II on day 5), 10% (G-II on day 10), and 23.33% (G-III on day 10) of total tested antimicrobials. The antimicrobials ampicillin, aztreonam, cefotaxime, ceftazidime, chloramphenicol, and tetracycline were converted from R → S; simultaneously more than 2 folds decreases in MIC values in lyophilized treated group G-III; cefotaxime showed a decrease in susceptibility from R → I in G-II on day 10. Besides, amoxicillin/K-clavulanate and ampicillin/sulbactam were converted from S → R in G-II and cefepime converted from I → S in G-III on day 10.
| S. No. | Antimicrobial | Control | Treated | ||
|---|---|---|---|---|---|
| G-I | G-II | G-III | |||
| Day-5 | Day-10 | Day-10 | |||
| 1 | Amikacin | R | R | R | R |
| 2 | Amoxicillin/k- clavulanate | S | S | R | S |
| 3 | Ampicillin/sulbactam | S | I | R | S |
| 4 | Ampicillin | R | R | R | S |
| 5 | Aztreonam | R | R | R | S |
| 6 | Cefazolin | I | I | I | I |
| 7 | Cefepime | I | I | I | S |
| 8 | Cefotaxime | R | R | I | S |
| 9 | Cefotetan | R | R | R | R |
| 10 | Cefoxitin | R | R | R | R |
| 11 | Ceftazidime | R | R | R | S |
| 12 | Ceftriaxone | S | S | S | S |
| 13 | Cefuroxime | R | R | R | R |
| 14 | Cephalothin | R | R | R | R |
| 15 | Chloramphenicol | R | R | R | S |
| 16 | Ciprofloxacin | S | S | S | S |
| 17 | Gatifloxacin | S | S | S | S |
| 18 | Gentamicin | I | I | I | I |
| 19 | Imipenem | S | S | S | S |
| 20 | Levofloxacin | S | S | S | S |
| 21 | Meropenem | S | S | S | S |
| 22 | Moxifloxacin | S | S | S | S |
| 23 | Nitrofurantoin | R | R | R | R |
| 24 | Norfloxacin | S | S | S | S |
| 25 | Piperacillin | S | S | S | S |
| 26 | Piperacillin/tazobactam | S | S | S | S |
| 27 | Tetracycline | R | R | R | S |
| 28 | Ticarcillin/k-clavulanate | S | S | S | S |
| 29 | Tobramycin | R | R | R | R |
| 30 | Trimethoprim/sulfamethoxazole | S | S | S | S |
Table 1: Effect of biofield treatment on Shigella sonnei to antimicrobial susceptibility pattern of selected antimicrobials
G, stands for group; The control group G-I and G-II were accessed on day 5 and 10; and G-III was accessed on day 10, after the biofield treatment; I, intermediate; S, susceptible; R, resistant.
| S. No. | Antimicrobial | Control | Treated | ||
|---|---|---|---|---|---|
| G-I | G-II | G-III | |||
| Day-5 | Day-10 | Day-10 | |||
| 1 | Amikacin | >32 | >32 | >32 | >32 |
| 2 | Amoxicillin/k- clavulanate | ≤8/4 | ≤8/4 | >16/8 | ≤8/4 |
| 3 | Ampicillin/sulbactam | ≤8/4 | 16/8 | >16/8 | ≤8/4 |
| 4 | Ampicillin | >16 | >16 | >16 | ≤8 |
| 5 | Aztreonam | >16 | >16 | >16 | ≤8 |
| 6 | Cefazolin | 16 | 16 | 16 | 16 |
| 7 | Cefepime | 16 | 16 | 16 | ≤8 |
| 8 | Cefotaxime | >32 | >32 | 32 | ≤16 |
| 9 | Cefotetan | >32 | >32 | >32 | >32 |
| 10 | Cefoxitin | >16 | >16 | >16 | >16 |
| 11 | Ceftazidime | >16 | >16 | >16 | ≤8 |
| 12 | Ceftriaxone | ≤8 | ≤8 | ≤8 | ≤8 |
| 13 | Cefuroxime | >16 | >16 | >16 | >16 |
| 14 | Cephalothin | 16 | 16 | 16 | 16 |
| 15 | Chloramphenicol | >16 | >16 | >16 | ≤8 |
| 16 | Ciprofloxacin | ≤1 | ≤1 | ≤1 | ≤1 |
| 17 | Gatifloxacin | ≤2 | ≤2 | ≤2 | ≤2 |
| 18 | Gentamicin | >8 | >8 | >8 | >8 |
| 19 | Imipenem | ≤4 | ≤4 | ≤4 | ≤4 |
| 20 | Levofloxacin | ≤2 | ≤2 | ≤2 | ≤2 |
| 21 | Meropenem | ≤4 | ≤4 | ≤4 | ≤4 |
| 22 | Moxifloxacin | ≤2 | ≤2 | ≤2 | ≤2 |
| 23 | Nitrofurantoin | >64 | >64 | >64 | >64 |
| 24 | Norfloxacin | ≤4 | ≤4 | ≤4 | ≤4 |
| 25 | Piperacillin | ≤16 | ≤16 | ≤16 | ≤16 |
| 26 | Piperacillin/tazobactam | ≤16 | ≤16 | ≤16 | ≤16 |
| 27 | Tetracycline | >8 | >8 | >8 | ≤4 |
| 28 | Ticarcillin/k-clavulanate | ≤16 | ≤16 | ≤16 | ≤16 |
| 29 | Tobramycin | >8 | >8 | >8 | >8 |
| 30 | Trimethoprim/sulfamethoxazole | ≤2/38 | ≤2/38 | ≤2/38 | ≤2/38 |
G, stands for group; MIC data is presented in μg/mL
Table 2: Effect of biofield treatment on Shigella sonnei to minimum inhibitory concentration (MIC) of selected antimicrobials.
Identification of S. sonnei by biochemical reactions
The results of biochemical reactions of S. sonnei are presented in Table 3, which represent a significant alteration in biochemical reactions of about 12.12% (G-II on day 5 and 10), and 57.58% (G-III on day 10) of total tested biochemicals as compared to control. The biochemicals such as adonitol, cephalothin, citrate, colistin, esculin hydrolysis, hydrogen sulfide, kanamycin, lysine, malonate, melibiose, raffinose, sorbitol, sucrose, tobramycin, urea, and Voges-Proskauer were changed from positive (control) → negative reactions (treated) in G-III microbes. Additionally, arginine was converted from positive to negative reaction in entire treated groups. Nitrofurantoin was converted from positive to negative in G-II on day 5 and G-III on day 10 (Table 3). Tartrate was converted from positive to negative reaction in both G-II and G-III on day 10; and inositol and tryptophan deaminase were converted from negative to positive reaction in G-II on both days (Table 3). All the data were compared as compared to control.
| S. No. | Code | Biochemical | Control | Treated | ||
|---|---|---|---|---|---|---|
| G-I | G-II | G-III | ||||
| Day-5 | Day-10 | Day-10 | ||||
| 1 | ACE | Acetamide | - | - | - | - |
| 2 | ADO | Adonitol | + | + | + | - |
| 3 | ARA | Arabinose | + | + | + | + |
| 4 | ARG | Arginine | + | - | - | - |
| 5 | CET | Cetrimide | - | - | - | - |
| 6 | CF8 | Cephalothin | + | + | + | - |
| 7 | CIT | Citrate | + | + | + | - |
| 8 | CL4 | Colistin | + | + | + | - |
| 9 | ESC | Esculin hydrolysis | + | + | + | - |
| 10 | FD64 | Nitrofurantoin | + | - | + | - |
| 11 | GLU | Glucose | + | + | + | + |
| 12 | H2S | Hydrogen sulfide | + | + | + | - |
| 13 | IND | Indole | - | - | - | - |
| 14 | INO | Inositol | - | + | + | - |
| 15 | K4 | Kanamycin | + | + | + | - |
| 16 | LYS | Lysine | + | + | + | - |
| 17 | MAL | Malonate | + | + | + | - |
| 18 | MEL | Melibiose | + | + | + | - |
| 19 | NIT | Nitrate | + | + | + | + |
| 20 | OF/G | Oxidation-fermentation/glucose | + | + | + | + |
| 21 | ONPG | Galactosidase | + | + | + | + |
| 22 | ORN | Ornithine | + | + | + | + |
| 23 | OXI | Oxidase | - | - | - | - |
| 24 | P4 | Penicillin | + | + | + | + |
| 25 | RAF | Raffinose | + | + | + | - |
| 26 | RHA | Rhamnose | + | + | + | + |
| 27 | SOR | Sorbitol | + | + | + | - |
| 28 | SUC | Sucrose | + | + | + | - |
| 29 | TAR | Tartrate | + | + | - | - |
| 30 | TDA | Tryptophan deaminase | - | + | + | - |
| 31 | TO4 | Tobramycin | + | + | + | - |
| 32 | URE | Urea | + | + | + | - |
| 33 | VP | Voges-Proskauer | + | + | + | - |
G, stands for group; -, (negative); +, (positive)
Table 3: Effect of biofield treatment on Shigella sonnie to biochemicals reactions pattern.
Effect of biofield treatment on biotype number
Biotype number of S. sonnie was determined on MicroScan Walk- Away® processed panel. The result was demonstrated an alteration in biotype number of S. sonnie in the entire treated groups G-II and G-III (Table 4). However, the species (S. sonnei) was remained unchanged in the entire treated group.
| Feature | Control | Treated | ||
|---|---|---|---|---|
| G-I | G-II | G-III | ||
| Day-5 | Day-10 | Day-10 | ||
| Biotype | 7736 7376 | 7776 5776 | 7776 5776 | 4300 1010 |
| Organism Identification Name | S. sonnei | S. sonnei | S. sonnei | S. sonnei |
Table 4: Effect of biofield treatment on Shigella sonnei to alteration in biotype.
16S rDNA gene sequencing
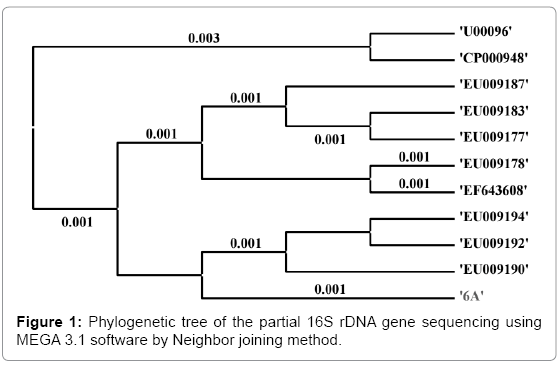
The 16S rDNA sequence was determined in S. sonnei and shown in Figure 1. The alignment and comparison of the gene sequences were performed with the sequences stored in Gene Bank data base available from NCBI using the algorithm BLASTn program. As evidenced from nucleotides homology and phylogenetic analysis the sample 6A (S. sonnei) was identified as the same species (S. sonnei) with 99% identity of gene sequencing data. Ten bacterial species and S. sonnei were considered as Operational Taxonomic Units (OTUs) to facilitate the investigation of phylogenetic relationship of S. sonnei among other related species. Total 1500 base nucleotide of 16S rDNA gene sequences were compared by multiple alignments with the help of ClustalW in MEGA3.1 [30], and the data are shown in Table 5. As evidenced from Table 6, the lowest value of genetic distance from S. sonnei was 0.002 base substitutions per site. The nearest homolog genus-species of S. sonnei (Genbank accession number: EU009190) was determined by analyzing the 16S rDNA sequencing and phylogenetic tree, and found to be Shigella flexneri (Genbank accession number: EF643608). The distance matrix was prepared based on nucleotide sequence homology data and presented in Table 6. All pairwise distance analysis was carried out employing the p-distance method in MEGA3.1 software.
| Alignment view | AN | Alignment results | Sequence description |
|---|---|---|---|
 |
6A | 0.99 | Sample studied |
 |
CP000948 | 0.99 | Escherichia colistrain DH10B, |
 |
U00096 | 0.99 | Escherichia colistrain K12 sub str. MG1655 |
 |
EU009194 | 0.99 | Shigellasonneistrain FBD020 |
 |
EU009192 | 0.99 | Shigellasonneistrain FBD018 |
 |
EU009190 | 0.99 | Shigellasonneistrain FBD016 |
 |
EU009187 | 0.99 | Shigellaflexneristrain FBD002 |
 |
EU009178 | 0.99 | Shigellaboydiistrain FBD007 |
 |
EF643608 | 0.99 | Shigellaflexneristrain FBD001shig |
 |
EU009183 | 0.99 | Shigelladysenteriaestrain FBD012 |
 |
EU009177 | 0.99 | Shigellaboydiistrain FBD006 |
AN: Accession Number
Table 5: The closest sequences of Shigella sonnei from sequence alignment using NCBI GenBank and Ribosomal database project (RDP).
| AN | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| EU009194 | 1 | — | 0.998 | 0.995 | 0.998 | 1.000 | 0.998 | 0.995 | 0.997 | 0.997 | 1.000 | 0.998 |
| EU009187 | 2 | 0.002 | — | 0.994 | 0.999 | 0.998 | 0.999 | 0.994 | 0.999 | 0.999 | 0.998 | 0.997 |
| U00096 | 3 | 0.005 | 0.006 | — | 0.994 | 0.995 | 0.994 | 1.000 | 0.993 | 0.993 | 0.995 | 0.994 |
| EU009178 | 4 | 0.002 | 0.001 | 0.006 | — | 0.998 | 0.999 | 0.994 | 0.998 | 0.998 | 0.998 | 0.997 |
| EU009192 | 5 | 0.000 | 0.002 | 0.005 | 0.002 | — | 0.998 | 0.995 | 0.997 | 0.997 | 1.000 | 0.998 |
| EF643608 | 6 | 0.002 | 0.001 | 0.006 | 0.001 | 0.002 | — | 0.994 | 0.998 | 0.998 | 0.998 | 0.997 |
| CP000948 | 7 | 0.005 | 0.006 | 0.000 | 0.006 | 0.005 | 0.006 | — | 0.993 | 0.993 | 0.995 | 0.994 |
| EU009183 | 8 | 0.003 | 0.001 | 0.007 | 0.002 | 0.003 | 0.002 | 0.007 | — | 1.000 | 0.997 | 0.997 |
| EU009177 | 9 | 0.003 | 0.001 | 0.007 | 0.002 | 0.003 | 0.002 | 0.007 | 0.000 | — | 0.997 | 0.997 |
| EU009190 | 10 | 0.000 | 0.002 | 0.005 | 0.002 | 0.000 | 0.002 | 0.005 | 0.003 | 0.003 | — | 0.998 |
| 6A | 11 | 0.002 | 0.003 | 0.006 | 0.003 | 0.002 | 0.003 | 0.006 | 0.003 | 0.003 | 0.002 | — |
AN: Accession Number
Table 6: Distance matrix based on nucleotide sequence homology (Using Kimura-2 Parameter).
Discussion
Antimicrobial resistance is a major global threat to public health, reported by World Health Organization (WHO). WHO also reported a post-antibiotic era, where people will die from simple microbial infections that have been curable for decades. Microbes naturally mutate and ultimately become immune to antimicrobials. Unfortunately, due to misuse of antimicrobials like over-prescribing by doctors or improper uses by patients is causing it to happen in much faster than expected. Similarly, S. sonnei has been acquired resistant to commonly used antimicrobials like tetracycline, streptomycin, trimethoprim, sulfonamide, and ampicillin. Further, emergence of extended-spectrum β-lactamases (ESBLs) was also detected in S. sonnei in some Asian countries like Korea [2,3,31].
Due to increasing the number of clinical specimens, costeffectiveness, and convenient interfaces with hospital information systems and laboratory the uses of automated or semi-automated systems for the susceptibility testing and identification of microbes has been increased recently [32]. Therefore, we also utilized the MicroScan Walk-Away® system for analysis of antimicrobial sensitivity, biochemical reactions, and biotyping. The overall result of antimicrobial susceptibility of biofield treated S. sonnei suggested that biofield treatment has significantly alerted the sensitivity of microbes in both side (either S → R or R → S) as compared to control. The biochemical reactions of treated cells of S. sonnei were altered in the range of 12.11 to 57.58% in treated group as compared to control, which could be due to some alteration happened in metabolic enzyme systems and/or genetic system. It was also found that there was an alteration of biotype number in treated groups of S. sonnei. Based on the BLASTn analysis, the sample 6A was identified as S. sonnei. The closest homologues species of S. sonnei was identified as Shigella flexneri. The present study revealed that biofield treatment can alter the sensitivity of antimicrobials against S. sonnei. It seems that biofield treatment can play a potential role to circumvent the severe microbial infection in the fast and cost effective way as compared to modern medication.
Conclusion
Altogether data suggest that there was an impact of biofield treatment on antimicrobial susceptibility, biochemical reactions pattern, and biotype number of S. sonnei. To the best of our knowledge, this is the first report describing the significant impact of biofield treatment on S. sonnei in relation to change the sensitivity of antimicrobials.
Acknowledgement
The authors would like to acknowledge the whole team of Hinduja Microbiology Lab for their support. The authors also like to acknowledge the Trivedi Science, Trivedi Master Wellness and Trivedi Testimonials for their steady support during the work.
References
- Haddad M (2014) Antimicrobial resistance of uropathogens and rationale for empirical therapy in Jordan. Biomed Pharmacol J 7: 1-8.
- Seol SY, Kim YT, Jeong YS, Oh JY, Kang HY, et al. (2006) Molecular characterization of antimicrobial resistance in Shigellasonnei isolates in Korea. J Med Microbiol 55: 871-877.
- Sur D, Ramamurthy T, Deen J, Bhattacharya SK (2004) Shigellosis : challenges & management issues. Indian J Med Res 120: 454-462.
- Abdu A, Aboderin AO, Elusiyan JB, Kolawole DO, Lamikanra A (2013) Serogroup distribution of Shigella in Ile-Ife, southwest Nigeria. Trop Gastroenterol 34: 164-169.
- Replogle ML, Fleming DW, Cieslak PR (2000) Emergence of antimicrobial-resistant shigellosis in Oregon. Clin Infect Dis 30: 515-519.
- Biswas S, Brunel JM, Dubus JC, Reynaud-Gaubert M, Rolain JM (2012) Colistin: an update on the antibiotic of the 21st century. Expert Rev Anti Infect Ther 10: 917-934.
- Lucchetti G, de Oliveira RF, Gonçalves JP, Ueda SM, Mimica LM, et al. (2013) Effect of Spiritist "passe" (Spiritual healing) on growth of bacterial cultures. Complement Ther Med 21: 627-632.
- Jain S, Mills PJ (2010) Biofield therapies: helpful or full of hype? A best evidence synthesis. Int J Behav Med 17: 1-16.
- Benor DJ (1990) Survey of spiritual healing research. Complement Med Res 4: 9-33.
- Rein G (1995) The in vitro effect of bioenergy on the conformational states of human DNA in aqueous solutions. AcupunctElectrother Res 20: 173-180.
- Trivedi MK, Tallapragada RM, Branton A, Trivedi D, Nayak G, et al. (2015) Potential impact of biofield treatment on atomic and physical characteristics of magnesium. Vitam Miner 3: 129.
- Dabhade VV, Tallapragada RR, Trivedi MK (2009) Effect of external energy on atomic, crystalline and powder characteristics of antimony and bismuth powders. Bull Mater Sci 32: 471-479.
- Trivedi MK, Tallapragada RR (2009) Effect of superconsciousness external energy on atomic, crystalline and powder characteristics of carbon allotrope powders. Mater Res Innov 13: 473-480.
- Trivedi MK, Patil S, Tallapragada RM (2012) Thought intervention through biofield changing metal powder characteristics experiments on powder characterisation at a PM plant. Future Control and Automation LNEE 173: 247-252.
- Trivedi MK, Patil S, Tallapragada RM (2013) Effect of biofield treatment on the physical and thermal characteristics of vanadium pentoxidepowders. J Material SciEng S11: 001.
- Trivedi MK, Patil S, Tallapragada RM (2013) Effect of bio field treatment on the physical and thermal characteristics of silicon, tin and lead powders. J Material SciEng 2: 125.
- Trivedi MK, Patil S, Tallapragada RMR (2015) Effect of biofield treatment on the physical and thermal characteristics of aluminium powders. IndEng Manage 4: 151.
- Shinde V, Sances F, Patil S, Spence A (2012) Impact of Biofield treatment on growth and yield of lettuce and tomato. Aust J Basic ApplSci 6: 100-105.
- Sances F, Flora E, Patil S, Spence A, Shinde V (2013) Impact of biofield treatment on ginseng and organic blueberry yield. Agrivita J AgricSci 35.
- Lenssen AW (2013) Biofield and fungicide seed treatment influences on soybean productivity, seed quality and weed community. Agriculture Journal 8: 138-143.
- Patil SA, Nayak GB, Barve SS, Tembe RP, Khan RR (2012) Impact of biofield treatment on growth and anatomical characteristics of Pogostemoncablin (Benth.). Biotechnology 11: 154-162.
- Nayak G, Altekar N (2015) Effect of biofield treatment on plant growth and adaptation. J Environ Health Sci 1: 1-9.
- Trivedi MK, Patil S (2008) Impact of an external energy on Staphylococcus epidermis [ATCC-13518] in relation to antibiotic susceptibility and biochemical reactions-an experimental study. J Accord Integr Med 4: 230-235.
- Trivedi MK, Patil S (2008) Impact of an external energy on Yersinia enterocolitica [ATCC-23715] in relation to antibiotic susceptibility and biochemical reactions: an experimental study. Internet J Alternat Med 6.
- Trivedi MK, Patil S, Shettigar H, Gangwar M, Jana S (2015) Antimicrobial sensitivity pattern of Pseudomonas fluorescens after biofield treatment. J Infect Dis Ther 3: 222.
- Fader RC, Weaver E, Fossett R, Toyras M, Vanderlaan J, et al. (2013) Multilaboratory study of the biomic automated well-reading instrument versus MicroScanWalkAway for reading MicroScan antimicrobial Susceptibility and identification panels. J ClinMicrobiol 51: 1548-1554.
- Gomaa FM, Tawakol WM, Abo El-Azm FI (2014) Phenotypic and genotypic detection of some antimicrobial resistance mechanisms among multidrug-resistant Acinetobacterbaumannii isolated from immunocompromised patients in Egypt. Egypt J Med Microbiol 23: 99-111.
- Jorgensen JH, Ferraro MJ (2009) Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 49: 1749-1755.
- Lennox VA, Ackerman VP (1984) Biochemical identification of bacteria by replicator methods on agar plates. Pathology 16: 434-440.
- Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5: 150-163.
- Cohen ML (1992) Epidemiology of drug resistance: implications for a post-antimicrobial era. Science 257: 1050-1055.
- Sader HS, Fritsche TR, Jones RN (2006) Accuracy of three automated systems (MicroScanWalkAway, VITEK, and VITEK 2) for susceptibility testing of Pseudomonas aeruginosa against five broad-spectrum beta-lactam agents. J ClinMicrobiol 44: 1101-1104.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 21702
- [From(publication date):
September-2015 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 17065
- PDF downloads : 4637