Research Article Open Access
Evaluation of Four Loop-Mediated Isothermal Amplification (LAMP) Assays for Identification of Shiga Toxin Producing E.Coli O157 (STEC) and Non-O157 Strains
James Mahony1,2*, Sylvia Chong2, Chris Stone2 and Linda Chui31Department of Pathology and Molecular Medicine, McMaster University, Canada
2Regional Virology Laboratory, St. Joseph’s Healthcare Hamilton, Hamilton, Canada
3Provincial Laboratory for Public Health, Walter Mackenzie Health Sciences Centre, University of Alberta Hospital, Edmonton, Alberta, Canada
- *Corresponding Author:
-
Mahony JB, Professor
Regional Virology Laboratory
St. Joseph’s Healthcare Hamilton
50 Charlton Ave. E, Hamilton
Ontario L8N 4A6,Canada
Tel: 905522-1155
Fax: 905521-6083
E-mail: mahonyj@mcmaster.ca
Received: March 01, 2016; Accepted: March 15, 2016; Published: March 21, 2016
Citation: Mahony J, Chong S, Stone C, Chui L (2016) Evaluation of Four Loop-Mediated Isothermal Amplification (LAMP) Assays for Identification of Shiga Toxin Producing E.Coli O157 (STEC) and Non-O157 Strains. Adv Mol Diag 1:104.doi:10.4172/amd.1000104
Copyright: © 2016 Mahony J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Advances in Molecular Diagnostics
Abstract
Shiga toxin-producing strains of E. coli are a significant cause of food-borne outbreaks of gastroenteric disease. The objective of this study was to evaluate loop-mediated isothermal amplification (LAMP) assays for the identification of Shiga toxin producing E. coli O157 (STEC) and non-O157 strains. Four LAMP assays were developed for the detection of the rfbE gene of E. coli O157, the Shiga toxin genes stx1/stx2 and eae intimin gene. The assays were run on a real time fluorometer (Genie II, Optigene, Horsham, UK) that displays real time amplification, the time to positivity and amplicon annealing temperature (Tm). The specificity of the LAMP assays was confirmed by testing a panel of 35 enteric bacteria, viruses and parasites, all of which tested negative in all four assays. The lower limit of detection for each of the gene targets was 10-100 genome equivalents and 1 cfu of E. coli O157. The LAMP assays were evaluated by testing a total of 135 stool specimens by LAMP, PCR or the xTAG® Gastrointestinal Pathogen Panel (GPP) assay (Luminex Molecular Diagnostics, Toronto, ON, Canada). Following resolution of the discordants and using positivity in two or more assays as the reference standard, the sensitivity of the LAMP assays was 100% (27/27), 98.4% (63/64) and 98.0% (47/48) for rfbE, stx1/stx2 and eae genes respectively, while the specificity for the assays was 100% (107/107), 100% (71/71) and 98.5% (66/67), respectively. The LAMP assays had excellent sensitivity and specificity for detecting Shiga toxin-producing E. coli O157 (STEC) and non-O157 in stool specimens and they were faster and more accurate than PCR. We suggest that these assays could be incorporated into E. coli O157 (STEC) testing algorithms.
Keywords
E. coli; Isothermal amplification; Shiga toxin
Introduction
Enterohemorrhagic Escherichia coli (EHEC) are food-borne pathogens that can cause serious illness, especially in young children and the elderly, and pose a serious global health concern [1,2]. E. coli O157 has been responsible for numerous food-borne outbreaks, and can result in hemorrhagic colitis or hemolytic uremic syndrome [3,4]. These bacteria are capable of producing large quantities of toxins (Shiga toxins) that can damage the intestinal lining and cause bloody diarrhea. More than 200 different serotypes of E. coli can produce Shiga toxin and at least 150 of these are human pathogens [5]. Shiga-toxin producing E. coli (STEC) strains cause approximately 176,000 illnesses, 2,400 hospitalizations, and 20 deaths annually in the USA [6]. Since surveillance for non-O157 STEC (O26, O45, O103, O111, O121, and O145) began in 2000, the incidence of non-O157 STEC infections surpassed that of O157 infections in the USA for the first time in 2010 [7,8]. Thus rapid and sensitive methods to detect both E. coli O157, as well as non-O157 serotypes, is required.
E. coli O157 and non-O157 STEC strains are usually detected using selective culture media, enzyme immunoassays or by commercial or in house PCR assays or by immunomagnetic separation (IMS) assays for food testing [9,10]. Immunomagnetic separation in combination with plating is the most common method used to detect E. coli O157 for food testing, but this approach is time-consuming and technically challenging. The recent outbreak of HUS in Germany attributed to a rare STEC serotype O104:H4 in sprouts adds additional demands on IMS testing [11]. In addition, the effective detection and isolation of non-O157 E. coli using traditional culture methods remains difficult. Currently many laboratories use enzyme immunoassays (EIAs) to test for O157 since only a few commercial assays or molecular tests have been approved by the FDA for the diagnosis of STEC infections. Nucleic acid amplification techniques such as PCR have been applied to detect E. coli O157 and non-O157 strains. Many PCR assays have been described for the detection of rfbE, stx1, stx2, eae and other genes present in STEC [12-14]. More recently, loop-mediated isothermal amplification (LAMP) has been applied to detect E. coli O157 in food, environmental water and human stools [15-20]. Our laboratory has recently shown that LAMP coupled with rapid specimen processing can detect respiratory viruses in nasal swabs in under 20 minutes [21,22]. More recently, other molecular epidemiological approaches including the detection of specific genetic loci as markers for STEC [23], and pulsed field gel electrophoresis, multilocus variable-number tandem repeat analysis (MLVA) and whole genome sequencing have been used to characterize E. coli isolates and to monitor outbreaks [24].
In this report, we evaluated four LAMP assays for the identification of STEC including O157 and non-O157 E. coli strains. The three assays detected four different E. coli genes, including the rfbE gene of E. coli O157, the Shiga toxin stx1/stx2 genes using a multiplex assay, and the eae virulence factor gene. The assays all had excellent sensitivity and specificity for detecting STEC and provide faster and more accurate results than PCR.
Materials and Methods
Clinical specimens and pre-analytical procedures
A total of 135 stool specimens submitted to the Regional Virology Laboratory at St. Josephs Healthcare Hamilton or to the Alberta Provincial Laboratory in Edmonton were used in the study. This study was approved by St. Joseph’s Healthcare Hamilton Research Ethics Review Board and the University of Alberta Ethics Review Board. In Hamilton, 20 stool specimens collected in March 2013 were processed as follows: 100-150 mg of bulk stool was added to SK38 bead tubes (Bertin Technologies, Montigny, France) containing Lysis Buffer (bioMérieux, St Laurent, Canada). This 10% stool suspension was vortexed for 5 minutes, allowed to stand at room temperature for 10 to 15 minutes, then centrifuged at 14,000 rpm for 2 minutes to pellet stool material, and 200 μL of the supernatant was used to extract total nucleic acid using the automated easyMag® (bioMerieux, St. Laurent, QC) as per the manufacturer’s Specific A protocol. An additional 115 stool specimens were collected at the Provincial Laboratory for Public Health (ProvLab) in Edmonton. Ninety of the 115 were first enriched by culture overnight in MacConkey broth and 200 μL was removed, centrifuged at 13,000 × g for 3 min, and the pellet was washed with 1 ml wash buffer consisting of 12 mM Tris buffer, pH 7.4. After re-centrifugation, the pellet was suspended in 200 μL rapid lysis buffer (100 mM NaCl, 10 mM Tris- HCL, pH 8.3, 1 mM EDTA, pH 9.0, 1% Triton X-100), boiled for 15 min and clarified by centrifugation at 13,000 × g for 15 min. A 1:10 dilution was performed and an aliquot (5 μL) of the diluted sample was subsequently used for molecular testing. An additional 25 isolates of known serotypes of STEC containing either stx1, stx2 or both stx1/stx2 genes from the ProvLab collection were also evaluated. These isolates were grown on BAP and a single colony was touched with a pipette tip, dispensed into 200 μL of rapid lysis buffer (above), boiled for 15 min and following centrifugation an aliquot (5 μL)of the supernatant was tested by PCR and LAMP.
E. coli O157 serotyping
E. coli serotyping was performed using O157 direct antibody agglutination (BD Difco Burlington, ON, Canada) and H7 antiserum (BD Difco) by tube flocculation as previously described [7,8].
PCR testing
Two conventional PCR assays were used in the study. One end point assay that amplifies 614 bp and 779 bp regions of the stx1 and stx2 genes, respectively was performed as described [12] A second endpoint PCR assay was used to resolve discordant results. This assay which amplifies a 259 bp region of the E. coli O157 rfb gene, 180 bp and 255 bp regions of the stx1 and stx2 genes, respectively, and a 384 bp region of the eae gene and was performed as described by Paton and Paton [13]. For all PCR assays 5 μL of extracted nucleic acid was used for each reaction.
xTAG® GPP Assay
Nucleic acid extracted from 20 stools collected at St. Joseph’s Healthcare Hamilton was tested in the xTAG® GPP Assay (Luminex Molecular Diagnostics, Toronto, ON, Canada) which detects 15 Gastrointestinal Pathogen targets including Campylobacter (C. jejuni, C. coli and C. lari only), Clostridium difficile toxin A/B, Cryptosporidium (C. parvum and C. hominis only), E. coli O157, Enterotoxigenic E. coli (ETEC) LT/ST, Giardia (G. lamblia only, also known as G. intestinalis and G. duodenalis), Norovirus GI/GII, Rotavirus A, Salmonella, Shiga Toxin-producing E. coli (STEC) stx1/stx2, and Shigella (S. boydii, S. sonnei, S. flexneri and S. dysenteriae). The xTAG® GPP assay was performed according to the manufacturer’s instructions using 10 μL of extracted nucleic acid from bulk stool prepared as described above.
Preparation of transcripts
PCR amplicons containing full length gene targets for LAMP viz. rfbE gene of E. coli O157, the stx1/stx2 genes and the eae gene were cloned into pGEM-T vector using standard methods. Transcripts were prepared using an in vitro transcription Kit (Ambion, Life Technologies, Burlington, ON) and RNA copy number was determined by reading absorbance at A260 nm (1 Absorbance unit equals 40 ug RNA).
LAMP assays
Four LAMP assays that detect either the E. coli O157 rfbE gene, the Shiga toxin genes stx1 and stx2, or the eae virulence factor gene were used to test for E. coli O157 (STEC) and non-O157 E. coli. The primers consisted of a set of 5 or 6 primers for each gene target and were purchased as RUO reagents from Canadian Molecular Developments (Division of Pro-Lab Diagnostics, Richmond Hill, ON, Canada). The final reaction volume for LAMP was 25 μL and consisted of 15 μL of ISO-0001 MasterMix (Optigene, Horsham, UK), 5 μL primer mix and 5 μL of either easyMag® extracted nucleic acid or boiled lysate as described above. The 5 μL primer mix for each gene target consisted of the following: F3 and B3 primers at 0.2 μM, FIP and BIP primers at 0.8 μM, and LF and LB primers at 0.4 μM as described previously. The specificity of each assay were determined by testing extracted nucleic acid from a range of other pathogens including both bacteria and viruses. The reactions were run on a real time fluorometer (Genie II from Optigene) at 65°C for 30-40 min, followed by heating and cooling steps of 98°C to 80°C (0.05°C/s) to allow re-annealing of any amplified DNA product. The Genie II instrument displays the amplification curve, the amplification time in min/sec and the annealing temperature of the amplified product. A positive result is indicated by either an amplification time (min and seconds), an amplification curve, or a melting temperature (Tm) within 2°C of the predicted Tm. A positive control consisting of E. coli O157 (EDL933) containing rfbE, stx1/ stx2, and eae genes was included in each run. All specimens were tested blindly by LAMP, PCR and the Luminex xTAG® GPP® assay. The sensitivity and specificity of the LAMP assays were calculated using a combined reference standard of positivity by two or more assays.
Results
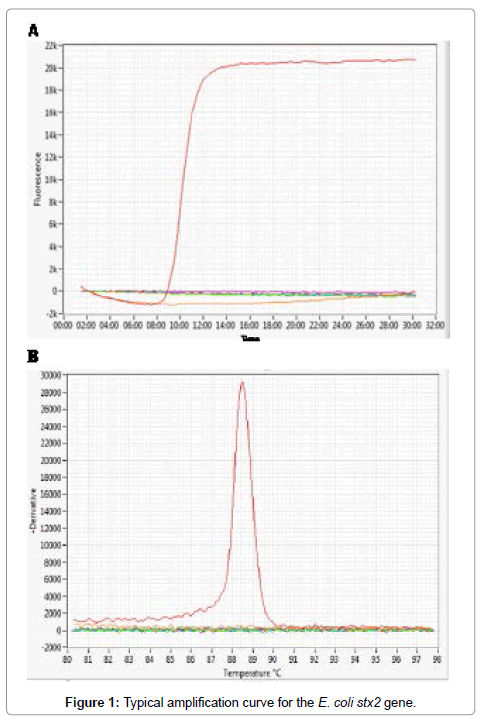
We evaluated four LAMP assays for the detection of Shiga toxinproducing E. coli O157 and non-O157 strains. Four sets of primers were used for the detection of four genes, including the rfbE gene of serotype O157 and three virulence factor genes viz. stx1,stx2 and eae. The LAMP assays for rfbE and eae were run as separate assays while the assays for stx1and stx2 were combined into a single multiplex LAMP assay. The assays were run on a real time fluorometer that displayed amplification signals in real time and at the end of the run displayed both the time to positivity and the annealing temperature (Tm) for each specimen that provided confirmation of a positive result. Figure 1 shows a typical amplification curve and Tm value for the stx2 gene. The Tm values for the four gene targets were between 83 and 88°C and were consistently within two degrees of the expected values for each target for all positive specimens. For the stx1/stx2 multiplex assay the Tm values for stx1 and stx2 were overlapping so that a positive result could not distinguish which of the two Shiga toxin genes was present. LAMP was successfully run on extracted nucleic acid and also on aliquots of cell lysates obtained using a rapid lysis method where a single colony was picked from an agar plate, boiled for 15 min, and 5 μL tested directly in the assay. Using this rapid method, we were able to identify the presence of all virulence genes in a single colony within 30-40 minutes.
To assess the specificity of the LAMP assays, we first tested purified nucleic acid from 35 different enteric specimens which represented 29 enteric bacteria, four enteric viruses viz. Adenovirus, Rotavirus and Norovirus GI and GII, and two parasites, Giardia and Cryptococcus. Nucleic acid was purified from S. aureus, E. faecalis, E. faecalis, M. luteus, S. saprophyticus, P. mirabilis, Y. entercolitica, S. Typhimurium,. S. marcescnes, S. sonnei, S. flexneri, K. pneumonia, P. vulgaris, P. aeruginosa, E. cloacae, S. boydii, ETEC LT/ST, E. coli O12826, Campylobacter, C. difficile, S. dysenteriae, Adenovirus, Norovirus, Rotavirus, Giardia and Crytosporidium. All extracted nucleic acid samples tested negative for all four LAMP targets (Table 1). E. coli O26:B6, E. coli O55:B5, E. coli O86: B7, E. coli O111:B4 and E. coli O121:B8 were negative for the O157 rfbE and stx1/stx2 genes but tested positive for the eae gene by LAMP. These results were confirmed by PCR using the primers described by Patton et al. [13]. Of note is the fact that specimens which typed as O rough:H7 and O22:H2 were positive by both LAMP and PCR for the rfbE gene using the Paton et al. E. coli O157 primers.
| Specimen ID | Amplification Time (mm:ss), Annealing Temperature (°C) | ||
|---|---|---|---|
| stx1/stx2 | eae | rfbE | |
| S. aureus (ATCC 25923) | - | - | - |
| S. epidermidis (ATCC 1228) | - | - | - |
| E. faecalis (ATCC 29212) | - | - | - |
| M. luteus (ATCC 49732) | - | - | - |
| S. saprophyticus(ATCC 15305) | - | - | - |
| P. mirabilis (ATCC 43071) | - | - | - |
| Y. entercolitica(ATCC 9610) | - | - | - |
| S. Typhimurium(ATCC 14028) | - | - | - |
| S. marcescens (ATCC 8100) | - | - | - |
| S. sonnei | - | - | - |
| S. flexneri (ATCC 12022) | - | - | - |
| K. pneumoniae (ATCC 13883) | - | - | - |
| P. vulgaris (ATCC 13315) | - | - | - |
| P. aeruginosa (ATCC 27853) | - | - | - |
| E. cloacae (ATCC 13047) | - | - | - |
| Shigellaboydii | - | - | - |
| ETEC LT/ST (clinical stool) | - | ND | - |
| Salmonella (clinical stool) | - | ND | - |
| E. coli (ATCC 25922) | - | ND | - |
| E. coli O26:B6* | - | 22:30, 83.40 | - |
| E. coli O55:B5* | - | 5.30, 83.67 | - |
| E. coli O86:B7* | - | 9.30, 83.80 | - |
| E. coli O128:B12 | - | - | - |
| E. coli O111:B4* | - | 18:30, 83.58 | - |
| E. coli O121:B8* | - | 16:15, 83.48 | - |
| Campylobacter (clinical stool) | - | ND | - |
| Shigella (clinical stool) | - | ND | - |
| C. difficile (clinical stool) | - | ND | - |
| Shigelladysenteriae | 16:15, 86.09 | - | - |
| Giardia (clinical stool) | - | ND | ND |
| Crytosporidium (clinical stool) | - | ND | - |
| Adenovirus (clinical stool) | - | ND | - |
| Norovirus GI (clinical stool) | - | ND | - |
| Norovirus GII (clinical stool) | - | ND | - |
| Rotavirus A (clinical stool) | - | ND | - |
Note:*E. coli O26:B6, E. coli O55:B5, E. coli O86:B7, E. coli O111:B4, E. coli O121:B8 also tested positive in the eae PCR from Paton et al. (1998). The E coli O157 LAMP assay detected samples typed to be Orough:H7 and O22:H2. But these samples also tested positive with the E. coli O157 primers from Paton et al. (1998).
Table 1: Specificity of the LAMP assays for E. coli stx1/stx2, eae and rfbE genes.
To determine the analytical sensitivity of the LAMP assays, we tested serial dilutions of in vitro transcripts prepared from cloned PCR products. All four LAMP assays had a lower limit of detection of between 10 and 100 genome equivalents (ge). The rfbE assay detected 10 ge in 2 out of 3 replicates, the stx1/stx2 assay detected 10 ge in 1/3 replicates while the stx2 and eae assays detected 100 ge in 2/3 and 3/3 replicates respectively (Table 2). All four LAMP assays were able to detect 10 CFU/mL (Table 3).
| Number of copies of rfbEtarget | Amplification Time (mm:ss), Annealing Temperature (°C) |
| 106 copies | 12:30, 85.47 |
| 105 copies | 13:45, 85.54 |
| 104 copies | 15:30, 85.82 |
| 103 copies | 17:00, 85.81 |
| 103 copies | 17:00, 85.34 |
| 103 copies | 17:00, 85.37 |
| 102 copies | 17.30, 85.60 |
| 102 copies | 22:00, 85.60 |
| 102 copies | 18:00, 85.59 |
| 10 copies | - |
| 10 copies | 17:45, 85.73 |
| 10 copies | 18:00, 85.84 |
| 1 copy | - |
| Number of copies of stx1 or stx2 target | Amplification Time (mm:ss), Annealing Temperature (°C) |
| Stx1 106 copies | 13:15, 85.79 |
| Stx1 105 copies | 16:00, 85.74 |
| Stx1 104 copies | 19:30, 85.83 |
| Stx1 103 copies | 21:30, 85.79 |
| Stx1 103 copies | * , 85.68 |
| Stx1 103 copies | 22:45, 85.44 |
| Stx1 102 copies | -, 85.99 |
| Stx1 102 copies | - |
| Stx1 102 copies | - |
| Stx1 10 copies | 29:00, 85.62 |
| Stx1 10 copies | - |
| Stx1 10 copies | - |
| Stx2 106 copies | 17:30, 86.99 |
| Stx2 105 copies | 18:45, 87.45 |
| Stx2 104 copies | 23:15, 87.40 |
| Stx2 103copies |
|
| Stx2 103 copies |
|
| Stx2 103 copies | - |
| Stx2 102 copies |
|
| Stx2 102 copies | *,87.27 |
| Stx2 102 copies | - |
| Stx2 10 copies | - |
| Stx2 10 copies | - |
| Stx2 10 copies | - |
| Number of copies of eaetarget | Amplification Time (mm:ss), Annealing Temperature (�°C) |
| 106copies | 11:00, 83.02 |
| 105 copies | 12:30, 83.01 |
| 104 copies | 13:30, 82.92 |
| 103 copies | 16:00, 82.53 |
| 103 copies | 14:45, 83.07 |
| 103 copies | 15:45, 83.17 |
| 102 copies | 15:15, 84.29 |
| 102 copies | 17:15, 82.92 |
| 102 copies | 17:15, 82.95 |
| 10 copies | - |
| 10 copies | - |
| 10 copies | - |
| 1 copy | - |
| 1 copy | - |
Note: The result is considered to be positive if either an amplification time and/or a Tm value within 2 degrees of the predicted Tm is recorded or if an amplification curve is displayed but the time to positivity is absent and the Tm is within range.
Table 2: Lower limit of detection of LAMP assays for stx1/stx2, eae and rfbE genes.
| Concentration of E. coli O157(EDL933) | Amplification Time (mm:ss), Annealing Temperature (°C) |
|---|---|
| 106 CFU/ml | 12:30, 84.19 |
| 105CFU/ml | 14:15, 84.18 |
| 104 CFU/ml | 16:15, 84.24 |
| 103 CFU/ml | 18:45, 84.20 |
| 102 CFU/ml | 20:15, 84.29 |
| 101 CFU/ml | 20:45, 84.18 |
Note: A stock culture of E. coli O157 (EDL 933) was diluted and aliquots containing 101-106 CFU were tested by LAMP using the rfbE primers. Similar results were obtained using the stx1, stx2, and eae LAMP assays.
Table 3: Lower limit of detection of rfbE LAMP assay for detecting E. coli O157.
We next evaluated the LAMP assays using stool specimens that tested positive for one or more enteric pathogens in the xTAG® GPP Assay (Table 4). Elevan of the 20 stool specimens were positive by either the rfbE or the stx1/stx2 LAMP assays. One specimen, GPP03-48B, tested positive for stx1/stx2 by the xTAG® GPP, but was negative by the LAMP stx1/stx2 assay. Sequencing of the xTAG® GPP amplfied product showed that this sample was negative for the stx1/stx 2 genes. A second specimen, GPP03-174B, was E. coli O157 positive in the xTAG®GPP test, but tested negative in the rfbE LAMP assay. Sequencing of the PCR amplicon showed the specimen to be E. coli O157 negative. After discordant resolution of these 20 specimens, the stx1/stx2 LAMP assay had a sensitivity of 100% (8/8) and specificity of 100% (12/12). Similarly, the rfbE LAMP assay had a sensitivity of 7/7 (100%) and specificity of 12/12 (100%). The stx1/stx2 and the rfbE LAMP results were negative for the stool specimens that were positive for Giardia, Adenovirus, Norovirus GI/GII, Rotavirus A, Campylobacter, ETEC LT/ST, C. difficile A/B, Cryptosporidium, Shigella, and Salmonella.
| Specimen ID | xTAG®GPP test result | LAMPbstx1/stx2 result (mm:ss,Tm) | LAMPbrfbE result (mm:ss,Tm) |
|---|---|---|---|
| S002 | E. coli O157, STEC stx1/stx2 | 15:15, 87.18 | 14:45, 84.09 |
| S010 | E. coli O157, STEC stx1/stx2 | 11:15, 87.27 | 11:00, 84.19 |
| S016 | E. coli O157, STEC stx1/stx2 | 15:00, 87.22 | 15:30, 84.04 |
| S028 | Campylobacter, STEC stx1/stx2 | 23:30, 86.78 | NA |
| S037 | E. coli O157, STEC stx1/stx2 | 15:00, 87.40 | 15:00, 83.85 |
| S092 | E. coli O157, STEC stx1/stx2 | 13:00, 87.40 | 12:45, 84.25 |
| GPP03-17B | Norovirus GI/GII, ETEC LT/ST, STEC stx1/stx2 | -*, 85.62 | - |
| GPP03-48B | Adenovirus 40/41, Rotavirus A, STEC stx1/stx2, Shigella | - | - |
| GPP03-90B | E. coli O157, Giardia, MS2 failure | - | 20:15, 84.27 |
| GPP03-25B | Adenovirus 40/41, Norovirus GI/GII, E. coli O157 | - | -, 84.29 |
| GPP03-174B | Rotavirus A, C. difficile toxin A/B, E. coli O157 | - | - |
| GPP03-198B | Rotavirus A, STEC stx1/stx2 | 9:45, 87.16 | - |
| GPP03-6B | Campylobacter, ETEC LT/ST | - | - |
| GPP03-10B | Norovirus GI/GII, Campylobacter, ETEC LT/ST, Salmonella | - | - |
| GPP03-11B | Campylobacter, Cryptosporidium | - | - |
| GPP03-14B | Adenovirus 40/41, Norovirus GI/GII | - | - |
| GPP03-156B | Rotavirus A, ETEC LT/ST, Salmonella | - | - |
| GPP03-160B | Rotavirus A, Salmonella | - | - |
| GPP03-170B | Rotavirus A, ETEC LT/ST, Salmonella | - | - |
| GPP03-15B | Shigella | - | - |
| E. coli O157 | 9:45, 87.16 | 9:30, 84.14 |
Note:aTwentyxTAG®GPPpositive stool specimens were tested by LAMP for the presence of stx1/stx2 and rfbE genes. LAMP results are expressed as time to positivity in mm:ss, Tm of the amplification curve. The result is considered to be positive if either an amplification time and/or a Tmvalue within 2 degrees of the predicted Tm is recorded or if an amplification curve is displayed but the time to positivity is absent and the Tm is within range.
Table 4: LAMP results for 20 xTAG®GPP positive stool specimens.
Next, we tested 25 samples from ProvLab’s collection which included 22 stx1/stx2 positive and where stx1/stx2 subtyping were performed. The stx1/stx2 LAMP assay correctly detected 21/22 positive specimens including the following stx subtypes stx1, 1a, 1c, 1d, 2a, 2b, 2c, 2d, 2e, and 2g for a sensitivity of 95.5% (21/22). One of the stx2f subtypes was missed (sample #24, Table 5). Three PCR negative specimens (#3, 4 and 6) were also negative by LAMP. Using PCR as the comparator, the LAMP eae and rfbE assays had a sensitivity of 100% (13/13) and 100% (7/7) and a specificity of 100% (12/12) and 100% (18/18), respectively.
| ID # | Serotypes (Stx status) | PCR Results | LAMP Results (mm:ss), Tm (°C) | |||
|---|---|---|---|---|---|---|
| rfbE | eae | stx1/stx2 | rfbE | eae | ||
| 1 | O111:H8 (Stx1/2) | - | + | 11:45, 87.42 | - | 11:45, 83.41 |
| 2 | O174:H8 (Stx1) | - | - | 13:00, 85.83 | - | - |
| 3 | Neg | - | - | - | - | - |
| 4 | Neg | - | - | - | - | - |
| 5 | O121:H11 (Stx2) | - | + | 15:45, 87.48 | - | 17:45, 83.69 |
| 6 | Neg | - | - | - | - | - |
| 7 | O25:H1 (Stx2) | - | + | 12:45, 87.55 | - | 10:30, 83.49 |
| 8 | O157:H7 (Stx1/2) | + | + | 12:15, 87.55 | 13:15, 84.32 | 11:15, 83.39 |
| 9 | O157:H7 (Stx1/2) | + | + | 13:00, 87.77 | 14:00, 84.24 | 11:45, 84.64 |
| 10 | O157:H7 (Stx1/2) | + | + | 12:15, 87.64 | 12:45, 84.21 | 10:30, 84.71 |
| 11 | O157:H8 (Stx1/2) | + | + | 15:45, 87.82 | 18:15, 84.43 | 15:45, 84.91 |
| 12 | O165:H25 (Stx1/2) | + | + | 12:45, 87.76 | 12:45, 84.46 | 10:45, 84.90 |
| 13 | ORough:H7 (Stx1/2) | + | + | 11:30, 87.68 | 12:00, 84.46 | 10:15, 84.84 |
| 14 | O26:H11 (Stx1) | - | + | 19:00, 86.08 | - | 18:45, 84.92 |
| 15 | Serotype ND (Stx1a) | + | + | 13:15, 87.42 | 12:30, 84.40 | 11:15, 83.22 |
| 16 | Serotype ND (Stx1c) | - | - | 14:15, 85.82 | - | - |
| 17 | Serotype ND (Stx1d) | - | - | 15:30, 85.53 | - | - |
| 18 | O103:H2 (Stx1) | - | + | 13:45, 85.59 | - | 18:00, 83.11 |
| 19 | Serotype ND (Stx2a) | - | - | 13:30, 85.92 | - | - |
| 20 | Serotype ND (Stx2b) | - | - | 24:00, 87.15 | - | - |
| 21 | Serotype ND (Stx2c) | - | - | 16:00, 87.54 | - | - |
| 22 | Serotype ND (Stx2d) | - | - | 17:00, 87.34 | - | - |
| 23 | Serotype ND (Stx2e) | - | - | 20:15, 87.46 | - | - |
| 24 | Serotype ND (Stx2f) | - | + | - | - | 15:15, 83.87 |
| 25 | Serotype ND (Stx2g) | - | - | 20:15, 87.39 | - | - |
Table 5: Comparison of PCR and LAMP results for 25 enriched stool specimens.
The LAMP assays were next evaluated by testing an additional 90 stools collected prospectively and submitted to the ProvLab testing for STEC testing in a research study. These were tested by routine enteric bacteria screening and by PCR for stx1/stx2. Eleven stools were positive for E. coli O157 by both conventional serotyping methods and the rfbE LAMP assay. Two additional stools were positive by rfbE LAMP assay but negative based on conventional serotyping method. These two discordants were also positive for the rfbE gene by PCR using the Paton primers. After discordant resolution, the sensitivity and specificity of the rfbE assay was 100% (13/13) and 100% (77/77) respectively. Thirty four of the stools were positive for either stx1 or stx2 using conventional PCR methods; 14 were positive for stx1, 5 were positive for stx2 and 15 were positive for stx1and stx2. All 34 PCR positives were positive by the stx1/stx2 LAMP assay. All 56 stx1 or stx2 negatives were also negative by the stx1/stx2 LAMP assay. The LAMP stx1/stx2 assay had a sensitivity and specificity of 100% (34/34) and 100% (56/56), respectively. Of the 34 stools that were positive for stx1 or stx2, there were 31 that tested positive by the eae LAMP assay. One of the eae LAMP negatives was negative by PCR using the Paton primers. There were four specimens that were negative by conventional methods that were positive by the LAMP eae assay, and three of these were also positive using the Paton PCR assay. Using a combined reference standard of positivity in two or more assays, the sensitivity and specificity of the eae LAMP assay for these 90 specimens was 97.1% (34/35) and 98.2% (54/55), respectively.
When the data was combined for all 134 stool specimens, the overall sensitivity and specificity for the rfbE LAMP assay was 100% (27/27) and 100% (107/107), respectively (Table 6). The overall sensitivity and specificity for the stx1/stx2 assay was 98.4% (63/64) and 100% (71/71) while the overall sensitivity and specificity for the eae assay was 98.0% (47/48) and 98.5% (66/67), respectively.
| LAMP assay* | % Sensitivity | % Specificity |
|---|---|---|
| rfbE | 100 (27/27) | 100(107/107) |
| stx1/stx2 | 98.4% (63/64) | 100 (71/71) |
| eae | 98.0 (47/48)* | 98.5 (66/67) |
Note: *A total of 135 stool specimens from two different study sites were tested by three LAMP assays.Only 134 specimens were tested by the rfbE primers as one specimen had insufficient volume for rfbE testing
Table 6: Overall performance of the LAMP assays for stool specimens.
Discussion
We evaluated four LAMP assays for detecting rfbE, stx1/stx2 and eae genes for the identification of Shiga toxin-producing E. coli O157 (STEC) and non-O157 E. coli. The LAMP assays for rfbE and eae were uniplex assays while the assay for the stx1/stx2 genes was a multiplex assay with primers for each gene. All assays were run on the Genie II real time fluorometer that displays the time to positivity and the annealing temperature of the product [21,22]. These assays could be used with either extracted nucleic acid from stool specimens or with overnight cultures (either broth or agar plate colonies) coupled with a rapid lysis method providing a rapid test result in under one hour. The time to positivity by LAMP was inversely correlated with the number of bacteria in the sample, increasing from 12.5 min for106 CFU /mL to 20.75 min for 1 CFU/mL (Table 3). All four LAMP assays had excellent analytical sensitivity with a lower limit of detection of 10 ge for rfbE and stx1/stx2 targets and 100 ge for the eae target. The rfbE assay was capable of detecting 1°C FU/mL. All LAMP assays had excellent specificity and gave negative results for stools that tested positive for other enteric pathogens (Table 1). We evaluated the performance of the three assays using a total of 135 stool specimens 45 of which were collected from two clinical studies and 90 were submitted for routine enteric bacteria screening. For processed stool specimens the sensitivity of the three assays was 100% (27/27) for rfbE, 98.4% (63/64) for stx1/ stx2 and 98.0% (47/48) for eae gene. The specificity of the three assays was also excellent; 100% (107/107) for rfbE, 100% (71/71) for stx1/stx2 and 98.5% (66/67) for eae. For cultured or processed stool specimens, including overnight culture enrichment followed by DNA extraction, the amplification times for the three gene targets ranged from 7 to 29 minutes allowing the LAMP assays to provide results in under an hour which was considerably faster than the 2-4 hours required for conventional PCR assays.
The majority of clinical microbiology laboratories today rely on culture-based techniques to identify E. coli O157. These include the use BBL CHROMagar (BD, Oakville, ON, Canada), which identifies O157 STEC based on a specific colony color or Sorbitol MacConkey agar plates which identify sorbitol non-fermenting E. coli O157. These culture based assays rely on phenotypic traits and typically require confirmation using molecular testing such as PCR. Testing stool specimens directly for Shiga toxin by either Vero cell culture requires cell culture expertise and can be time consuming. The ImmunoCardSTAT! Test from Meridian Bioscience (Cinncinati, OH, USA) test can be performed directly on stools and gives results in about 30 minutes but the sensitivity is poor. A new lateral flow test called the QUIK CHEK assay has an improved sensitivity around 80% but both tests miss a significant number of E. coli STEC positives [8,25]. Many different PCR assays have been developed for detecting E. coli O157. Although these assays show differences in performance, PCR generally has good sensitivity and high negative predictive values compared to culture and provide results much faster than the 22-30 hours required for culture. In one study, detecting E. coli O157 or non-O157 STEC by PCR had a lower specificity and sensitivity compared with conventional methods especially when targeting the stx2 gene which was due to genetic polymorphisms [9]. Recently, we proposed an algorithm for STEC screening and isolation using several well-characterized techniques including routine stool culturing, the QUIK CHEK assay and real-time PCR [10]. The QUIK CHEK assay alone was not sufficient, but could be combined with PCR to achieve acceptable sensitivity and specificity. If either the QUIK CHEK assay or real-time PCR was positive, further isolation was recommended using BBLTM CHROMagar O157 and Colorex STEC plates, followed by confirmation with PCR. The LAMP assays described in this study had excellent sensitivity and specificity for detecting E. coli O157 or non-O157 STEC and represent an improvement over PCR in both accuracy and provide a faster turn-around time. All the LAMP assays provided results in less than one hour compared with conventional PCR assays requiring 2-4 hours or real-time qPCR assays that can provide results in 45 minutes [25]. We show here the identification of E. coli STEC in stool specimens using LAMP assays without the need for screening with immunoassays or enrichment by overnight culture. These LAMP assays could be included in a testing algorithm as either front line testing of immunoassay screen positive stool specimens or for confirmatory testing of E. coli O157 and non-O157 following culture enrichment. We do not know the performance of these assays when used directly on stool specimens that have not been enriched by overnight culture. This is the subject of an ongoing study.
Acknowledgements
The authors acknowledge the secretarial assistance of Cathy McIntyre.
Conflict of Interest
No conflict of interest
References
- Currie A, MacDonald J, Ellis A, Siushansian J, Chui L, et al. (2007)Outbreak of Escherichia coli O157:H7 infections associated with consumption of beef donair. J Food Prot 70: 1483-1488.
- Denny J, Bhat M, Eckmann K (2008) Outbreak of Esherichia coli O157:H7 associated with raw milk consumption in the Pacific Northwest.Foodborne Pathog Dis 5: 321-328.
- Nataro JP, Kaper JB (1998) Diarrheagenic Escherichia coli. Clin Microbiol Rev 11: 142-201.
- Karmali M, Petric M, Lim C, Flemming PC, Arbus GS, et al. (1985) The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J Infect Dis 189: 556-563.
- Johnson KE, Thorpe CM, Sears CL (2006) The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli.Clin Infect Dis 43: 1587-1595.
- http://t.perishablepundit.com/docs/foodborneillness.pdfBrooks JT (2005) Non-O157 Shiga toxin-producing Eschericia coli infections in the United States, 1983-2002. J Infect Dis 192: 1422-1429.
- U.S. Department of Agriculture (2011) Draft risk profile of pathogenic non-O157 Shiga toxin-producing Escherichia coli. U.S. Department of Agriculture, Washington DC.
- Chui L, Couturier M, Chiu T, Wang G, Antonishyn N, et al. (2010) Comparison of shiga toxin-producing Escherichia coli detection methods using clinical stool samples. J MolecDiag 12: 469-475.
- Chui L, Lee M-G, Allen R, Bryks A, Haines L, et al. (2013) Comparison between ImmunCard STAT!® and real-time PCR as screening tools for both O157:H7 and non-O157 Shiga toxin-producing Escherichia coli in Southern Alberta, Canada. Diagn Microbiol Infect Dis; 77: 8-13.
- Buchholz U (2011) German outbreak of Escherichia coli O104:H4 associated with sprouts.N Engl J Med 365: 1763-1770.
- Gannon VP, King RK, Kim JY, Thomas EJ (1992) Rapid and sensitive method for detection of Shiga toxin-producting Escherichia coli in ground beef using polymerase chain reaction. Appl Environ Microbiol 58: 3809-3815.
- Paton AW, Paton JC (1998) Detection and characterization of Shiga toxin Escherichia coli by using the multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E coli hlyA, rfb0111, and rfb0157. J. Clin. Microbiol 36: 598-602.
- Wasilenko J, Fratamico P, Narang N, Tillman G, Ladely S (2012) Influence of primer sequences and DNA extraction method on detection of non-O157 shiga toxin-producing Escherichia coli in ground beef by real-time PCR targeting the eae, stx and serogroup-specific genes. J Food Protect 75: 1939-1950.
- Maruyama F, Kenzaka T, Yamaguchi N, Tani K, Nasu M (2003) Detection of bacteria carrying the stx2 gene by in situ loop-mediated isothermal amplification. Appl Environ Microbiol 69: 5023-5028.
- Hill J, Beriwal S, Chandra I, Paul V, Kapil A, et al. (2008) Loop-mediated isothermal amplification assay for rapid detection of common strains of Escherichia coli. J Clin Microbiol 46: 2800-2804.
- Zhao X, Li Y, You L, Xu Z, Li L, et al. (2010) Development and application of a loop-mediated isothermal amplification method on rapid detection Escherichia coli O157 strains from food samples. 37: 2183-2188.
- Wang F, Jiang L, Ge B (2011) Loop-mediated isothermal amplification assays for detecting shiga toxin-producing Escherichia coli in ground beef and human stools. Appl Environ Microbiol 91-97
- Wang F, Jiang L, Prinyawinwatul W, Ge B (2012) Rapid and specific detection of Escherichia coli serogroupds O26, O45, O103, O111, O121, O145 and O157 in ground beef, beef trim, and produce by loop-mediated isothermal amplification. Appl Environ Microbiol 2727-2736.
- Dong HJ, Cho AR, Hahn TW, Cho S (2014) Development of a multiplex loop-mediated isothermal amplification assay to detect Shiga toxin-producing Escherichia coli in cattle.J Vet Sci 15: 317-325.
- Mahony J, Chong S, Bulir D, Ruyter A, Mwawasi K, et al. (2013) Development of a sensitive loop-mediated isothermal amplification (LAMP) assay providing specimen-to-result diagnosis of RSV infections in 30 minutes. J Clin Microbiol 51: 2696-2701.
- Mahony J, Chong S, Bulir D, Ruyter A, Mwawasi K, et al. (2013) Multiplex loop-mediated isothermal amplification (M-LAMP) for the detection of Influenza A/H1, A/H3 and B can provide a specimen-to-result diagnosis in fourty minutes with a single genome copy sensitivity. J Clin Virol58: 127-131.
- Chui L, Li V, Fach P, Delannoy S, Malejczyk K, et al. (2015) Patterson-Fortin L, Poon A, King R, Simmonds K, Scott A, Lee M-C. Molecular profiling of Escherichia coli O157:h7 and Non-O157 strains isolated from humans and cattle in Alberta, Canada. J Clin Microbiol 53: 986-990.
- Berenger BM, Berry C, Peterson T, Fach P, Delannoy S, et al. (2015) The utility of multiple molecular methods including whole genome sequencing as tools to differentiate E. coli O157:H7 outbreaks.Euro Surveill 20: 47.
- Chui LL, Patterson Fortin J, Kuo V, LiV (2015) Boras Evaluation of Enzyme Immunoassays and Real-time PCR for detecting Shiga toxin-producing Escherichia coli in Southern Alberta, Canada. J Clin Microbiol.53: 1019-1023.
- Chui L, Patterson-Fortin L, Kuo J, Li V, Boras V (2015) Evaluation of Enzyme Immunoassays and Real-time PCR for detecting Shiga toxin-producing Escherichia coli in Southern Alberta, Canada. J Clin Microbiol 53(3): 1019-23.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 12432
- [From(publication date):
April-2016 - Apr 03, 2025] - Breakdown by view type
- HTML page views : 11461
- PDF downloads : 971