Research Article Open Access
Establishment and Characterization of a Novel Kidney-cell Line from Orange-spotted Grouper, Epinephelus coioides, and its Susceptibility to Grouper Iridovirus
Sue-Min Huang1,2, Chien Tu1, Shu-Ting Kuo1, Hung-Chih Kuo3, Chi-Chung Chou4 and Shao-Kuang Chang2*
1Animal Health Research Institute, Council of Agriculture, Taipei, Taiwan
2Institute of Veterinary Medicine, School of Veterinary Medicine, National Taiwan University, Taipei, Taiwan
3Department of Veterinary Medicine, National Chiayi University, Chiayi, Taiwan
4Department of Veterinary Medicine, National Chung-Hsing University, Taichung, Taiwan
- *Corresponding Author:
- Chang SK
Institute of Veterinary Medicine, School of Veterinary Medicine
National Taiwan University, Taipei 10617, Taiwan
Tel: +886- 2-3366-3864
E-mail: changsk@ntu.edu.tw
Received date: April 07, 2016; Accepted date: May 23, 2016; Published date: May 31, 2016
Citation: Huang SM, Tu C, Kuo ST, Kuo HC, Chou CC, et al. (2016) Establishment and Characterization of a Novel Kidney-cell Line from Orange-spotted Grouper, Epinephelus coioides, and its Susceptibility to Grouper Iridovirus. J Marine Sci Res Dev 6:192. doi:10.4172/2155-9910.1000192
Copyright: © 2016 Huang SM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Marine Science: Research & Development
Abstract
A new continuous cell line, designated as GK-7, was developed from the kidney tissue of the marine grouper, Epinephelus coioides. The cell line grew well in Leibovitz’s L-15 medium supplemented with 15% fetal bovine serum at a range of temperatures from 20 to 32â�?�?, with optimal growth at 25â�?�?. Morphologically, the GK-7 cell line is spindleshaped epithelial-like cell comfirmed by immunophenotyping with cytokeratin antibody. Chromosome number analysis showed that GK-7 cells of the 50th and 150th cell passages had a modal diploid chromosome number of 48 and 66, respectively. Replication of GIV with the cell line showed that the maximum virus yield reached up 108.4 TCID50 mL-1 with the cells of the 50th passage. Electron micrographs showed abundant cytoplasmic icosahedral virions with a mean diameter of 200 nm in virus-infected cells. Negative staining of ultrathin sections of infected cells showed three-layered membrane enveloped mature viral particles with a diameter about 240 nm. Green fluorescent protein can be expressed in both cell lines at 48 hr after the cell lines were transfected with a green fluorescent reporter gene driven by a cytomegalovirus promoter. Our results showed that the GK-7 cell line provided valuable tools for the isolation and investigation of fish iridovirus and for vaccine production.
Keywords
Fish cell line; Epinephelus coioides; iridovirus correspondence
Introduction
Grouper, Epinephelus spp., is one of the most important maricultured fish with great economic value, especially in south-east Asian countries. In Taiwan, the annual cultured grouper production is around 4,500 tons [1-6]. In the last 10 years, iridovirus and nervous necrosis virus (NNV) impacted the grouper industry severely and caused heavy economic losses. Developing an ideal cell line for in vitro studies of cell-virus interactions, virus propagation, isolation and vaccine development is critical to disease control in aquaculture. Taiwanese iridovirus isolates consist of three major genotypes : grouper iridovirus (GIV) which belong to Ranavirus, red sea bream iridovirus (RSIV), and infectious spleen and kidney necrosis virus (ISKNV) that are classified to Megalocystivirus genuses [7]. Hence, development a continuous cell line from tropical fish species for identifying pathogenesis of viral diseases and for vaccine production is imperative. To date, more than 22 cell lines have been developed from grouper for iridovirus and NNV isolation, such as seven cell lines established from eye, fin, heart, swimbladder, kidney, liver and brain of E. awoara; four from brain, fin, eye and spleen of E. coioides [3,8-23]; five from snout, spleen, live, kidney and swimbladder of E. akaara [4,9,15,24,25]; three from brain, gill, heart of E. quoyanus [10]; one from heart of E. lanceolatus [5]; one from embryo cells of E. tauvina [17]; and one from fin of E. fuscoguttatus [22]. Although four cell lines derived from E. coioides, have been documented, no cell line has been derived from kidney of the species. Fish spleen and kidney are important organs for virus infection, particularly for grouper iridovirus infection [6]. Availability of the cell lines from these tissues would be helpful for providing essential information on virus pathology and vaccine development studies. In this study, we established and characterized a new cell line from kidney tissue of orange spotted grouper. Susceptibility of a new cell line to GIV was confirmed by cytopathic effects (CPE) observation, virus titer determination, electron microscopy and comparison the infection kinetics of GIV replication in earlier and later cell passages in the routine culture system.
Materials and Methods
Primary cell culture
Healthy orange spotted grouper weighing 250 g were obtained from fish farm and maintained in laboratory aquaria equipped with seawater recirculation for 7 days. The fish were anesthetized with an overdose of MS-222 (Sigma-Aldrich, St. Louis, MO) and swabbed with 70% ethanol. The spleen, gill, kidney and fin were immediately removed and washed sequentially in phosphate-buffered saline (PBS) containing 5% chlorex for 10 secs, in 0.1% iodine solution for 2 min, and in antibiotic- antimycotic solution (Gibco, Grand Island, NY) for 1 hour. After washing, the tissues were minced with scissors and transferred to a 60 mm diameter tissue culture dish containing 10 ml of 0.25% trypsin-EDTA solution (Gibco) with a magnetic stirrer for gently digesting and then filtered through a 100 μm nylon mesh. After centrifugation at 1200 rpm for 15 min at 4�??, the filtered cell pellet was resuspended in Leibovitz’s L-15 (Life Technologics, Carlsbad, CA) cell culture medium containing 20% fetal bovine serum (FBS, Gibco), 200 IU mL-1 penicillin and 200 μg mL-1 streptomycin. The cells were seeded into 25 cm2 tissue culture flasks and incubated at 25�??.
Subculture maintenance and cryopreservation
When the primary cell cultures were grown to a complete monolayer, cells were washed with 0.25% trypsin solution and subcultured in fresh L-15 medium containing 20% FBS, 200 IU ml-1 penicillin and 200 μg mL-1 streptomycin. For the first 10 subcultures of the primary cells, the 50% culture medium was replaced with fresh medium. After the 10 passages, the concentration of FBS was decreased from 20% to 10% for the following 50 subcultures and the antibiotics were reduced to normal concentrations (100 IU ml-1 penicillin and 100 μg mL-1 streptomycin, Gibco). The subcultures were cultured in the growth medium at a split ration of 1:3 until 150 passages for the experiments. The subcultures were stored in liquid nitrogen after every 10th passage in the freezing medium, which consisted of FBS containing 10% dimethyl sulphoxide. Briefly, 2-day-old subcultures of the cells were harvested by centrifugation and suspended in the freezing medium. The cell suspension were transferred to a cryovial (Nunc) and kept at -80�?? for 48 hrs, and then stored in liquid nitrogen (-196�??). For revival, the cryovial was thawed quickly in a water bath at 25�?? and removal of the freezing medium by centrifugation, then the thawed cells were transferred to a 25-cm2 cell culture flasks with 5 ml culture medium. The viability was tested by a haemocytometer counting after trypan blue staining.
Effect of temperature and FBS concentration on cell growth
Optimal temperature and FBS concentrations for the E. coioides kidney (designated as GK-7) cell line was studied at the 50 passages. Briefly, cells were seeded onto 25-cm2 cell culture flasks with 1 × 105 cells per flask and incubated at 25�?? for more than 2 h to allow the cell to attach. The flasks with the attached cells were incubated at 20, 25, 28 and 32�?? and observed on a daily basis. Similar procedures were performed for determining the effects of various concentrations of FBS (2%, 5%, 10% and 15% FBS ) at 25�??. According to the above results, we also investigated the cell growth in L-15 containing 10% FBS at 25�?? in 50th and 150th different passages. Cells from each flasks were trypsinized, and counted by three independent hemocytometer counts every three days. The experiments were carried out for 15 days. The mean number of cells and standard deviations (SD) were calculated.
Chromosome number analysis
Chromosome numbers of the GK-7 cells were determined at 50th and 150th passages. The cells were incubated in a 25-cm2 tissue culture flask until 70% confluent and then treated with 0.2 μg ml-1 colcemid (Sigma) for 5 h at 25�??, allowing the cells arrested at metaphase. After gentle pipetting, the cells were collected by centrifugation at 200 g for 5 min at 4�?? and resuspended in hypotonic 0.75% KCl solution for 30 min. The resuspended cells were then fixed in a fixed solution (3:1 mixture of methanol : acetic acid) for 20 min at room temperature. Following centrifugation at 800 g for 10 min at 4�??, the supernatant was discarded and the cells were fixed in fresh fixed solution and centrifuged. The step was repeated the cell suspension and dropped onto a clean slide glass. After air-dry, the dried cells were stained with 5% Giemsa solution for 20 min at room temperature. One hundred cells at metaphase were counted under a light microscope (Nikon).
Virus
The GIV/90/grouper strain was used in this research. The virus strain was isolated from sick juveniles of grouper (Epinephelus coioides) in the field. Our data of viral major capsid protein (MCP) nucleotide phylogenetic analysis showed that the GIV/90/grouper isolates (GenBank accession number KU510328) belong to the GIV (Accession number AAV91066) group of Ranavirus. The MCP nucleotide sequence of GIV/90/grouper strain was 100% identical to that of GIV.
Confirmation of the origin of the cell line
To authenticate the origin of the developed GK-7 cell line, the complete sequence of the 16S ribosomal RNA (rRNA) was determined. Total genomic DNA was extracted from the 50th passage for GK-7 cells with a QIAamp DNA Mini kit (QIAGEN) according to the manufacturer’s instructions. Primers 16s-f (5’-taccgactcgctagccca-3’) and 16s-r (5’-tagtcgttttctcagaca-3’) were used to amplify a fragment of 16S rRNA gene that referred to the complete genome of mitochondrion derived from E. coioides (Accession number KM377093) to design primers. The polymerase chain reaction (PCR) was carried out in a 25 μl of mixtures containing 1x PCR buffer for Blend Taq (Toyobo), 0.2 mM dNTPs, 10 pmol of each primer, 1.25 unit of Blend Taq DNA polymerase, 100 ng of the extracted genomic DNA. The PCR mixture was incubated in a PTC-200 DNA thermal cycler (MJ Research, Waltham, MA) at 95�?? for 5 min of 1 cycle; 35 cycles of 94�?? for 30 s, 58�?? for 50 s, and 72�?? for 1 min ; finally, extension at 72�?? for 7 min. PCR products were sequenced using an ABI PRISM 377 DNA sequencer with a BigDye Terminator Kit (Applied Biosystems, Foster city, CA, USA). Sequences of the mitochondrial genes amplified from the GK-7 cell line were examined for similarity with existing sequences submitted in Gene Bank data base.
Morphological confirmation with immunocytochemistry
Immunophenotyping of the GK-7 cell line was carried out at passage 48 following the method described by Gong et al. Briefly, the cells were grown on coverslips in 6-well plates for 24 h and then fixed with 4% paraformaldehyde at 4�?? for 1 h and permeabilized with 0.2% Triton-X 100 at room temperature for 15 min. After blocking with 2% BSA in PBS at room temperature for 2 h, the cells were incubated with four different primary antibodies against mouse anticytokeratin, mouse antivimentin, mouse antifibronectin and goat anit rabbit desmin, respectively (Sigma-Aldrich; catalogue number: C2562, 180- 0052, F0791 and D8281), at a dilution of 1:200 at room temperature for 2 h to characterize the cell line. Subsequently, the cells were rinsed three times with PBS and were incubated with goat-anti-mouse IgG FITC-conjugated or goat-anti-rabbit IgG FITC-conjugated (1:320 in PBS containing 1% BSA). In control coverslips, only PBS with 1% BSA was used in place of the primary antibodies. DAPI (diaminophenylindole) at a final concentration of 1 μgmL-1 was used to stain DNA in nuclei. All samples were observed with a fluorescence microscope.
GIV infection, propagation and titration
For the virus isolation, the tissues were thawed and homogenized with 10-fold volume of sterile PBS (pH 7.4). The homogenate was centrifuged at 1500 g for 15 min at 4�?? and filtered through 0.45 μm membrane filter. The GK-7 cells were seeded onto 6-well plates at about 80% confluence and cultivated for 18 h; then, the medium was discarded and the GK-7 cells in the 6-well plates were inoculated with 0.1 mL of the filtrate. These inoculated cells were incubated at 25�?? for 7-14 days and observed daily by inverted microscope. After the 7-14 days, when CPE was not observed, supernatant were treated by the second to fifth passages with the same procedures. Virus titres were determinated by titrating a series of 10-fold dilutions onto each cells in 96-well plates and calculated as TCID50 mL-1 per flask [20].
Comparison of viral replication efficiency in earlier and later passages
The GIV replication in the GK-7 cell line was evaluated by virus inoculation in the 50th and 150th passages of the GK-7 cell line. Each cell passages were incubated in 25-cm2 tissue culture flasks at a density of 2 × 105 cells for 24 h at 25�?? to 80% confluence. After removal of the medium, the cell culture was inoculated with 0.1 mL of virus suspension at a multiplicity of infection (MOI) of 0.01, then 5 mL of maintenance medium containing 2% FBS was added. The cells were examined and their culture supernatants were sampled daily for 7 days after the inoculation. The sampled supernatants were stored at -80°C before determining virus titers and titrated in triplicate.
Electron microscopy
The monolayers of GK-7 cells were infected with 106 TCID50 mL-1 GIV/90/grouper. After appearance of advanced CPE, the virus-infected cells were harvested, pelleted by centrifugation at 3000 rpm for 10 min, and fixed with 2.5% glutaraldehyde in cacodylate buffer (0.1 M, pH 7.2) for 2 h at 4�??. The fixed cells were washed with fresh cacodylate buffer and rinsed in PBS buffer (0.1 M, pH 7.2) for 10 min three times,then post-fixed in 1% osmium tetroxide for 1 h. After being rinsed four times in PBS for 15 min, the fixed cells were dehydrated in graded ethyl alcohol (50, 75, 90, 95, 100%) and embedded in epoxy resin. Ultrathin sections were cut with a ultramicrotome (Reicher-Jung) and were then stained with saturated aqueous uranyl acetate and lead citrate. Preparation of GIV for negative staining were performed as described previously [18]. A drop of the virus infected cells which were treated by three cycles of rapid freezing/thawing were applied onto a carbon coated 300-mesh copper grids and stained with 2% phosphotungstic acid (pH 6.8) for 1 min. Specimens were examined using an electron microscope (Jeol JEM-1000CX II, Japan) and photomicrographs were taken.
Cell transfection
To evaluate transfection efficiency and gene expression in GK-7 cells, the vector pEGFP-N1expressing green fluorescent protein by human cytomegalovirus promoter was prepared according to supplier’s instructions (Clontech Laboratories, CA, USA). The cell line was seeded onto 24-well plates at a density of 2 × 104 cells per well. After 24 h, subconfluent monolayers were formed and the monolayers were washed once with serum-free mediun. Two micrograms of the plasmid pEGFP-N1 and 12 μL of lipofectamine 2000 (Invitrogen) were dissolved separately in 375 μL of L-15 medium without serum and antibiotics, and then they were mixed and incubated for 20 min to prepare the transfection reagent. Following the incubation, 500 μL of this mixture was added to each well. After 24-72 h, the green fluorescence signals were observed under a fluorescent microscope.
Results
Primary cell culture and subculture
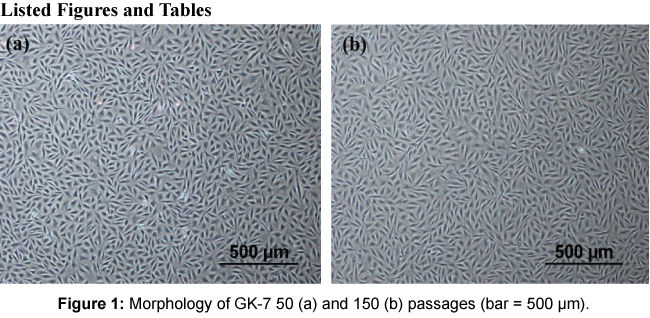
Primary cell cultures of E. coioides were carried out in November 2007 using tissues from spleen, gill, kidney and fin. The primary cells attached to the culture flask at day 2 and a monolayer of the cells was formed after 30 days. The cells were subcultured in L-15 medium with 20% FBS at a ratio of 1:2 every 5-7 days for the initial 10 passages, as allowed by the cell growth rates. After the 10 passages, these cells propagated rapidly and thereafter the cultures were adapted to culture medium with 10% FBS at 1:3-1:5 ratio at intervals of 3 to 5 days. After the 50th passage, the cells were split at a ratio of 1:3 every 7 days. Only the cells derived from kidney tissue formed a continuous cell line designated as GK-7. The passage numbers of the cells used in the present study were all around the 50th passage to 150th passage. Morphologically, the initial subcultures of GK-7 cell lines consisted of spindle-shape epithelial-like cells. To date, the cells have undergone 150 passages and showed no apparent morphologyical changes (Figure 1).
Cryopreservation
The GK-7 cells were cryopreserved every ten passages from 5th to 150 th passages. The cryopreserved cells at 15, 25, 45, 55, 65, 100 and 150 passages recovered from liquid nitrogen storage after 6 mo, 1 year, 2 years, 3 years showed 70-90% viability and grew to confluency within 3-5 days in L-15 medium with 10% FBS at 25�??.
Growth studies
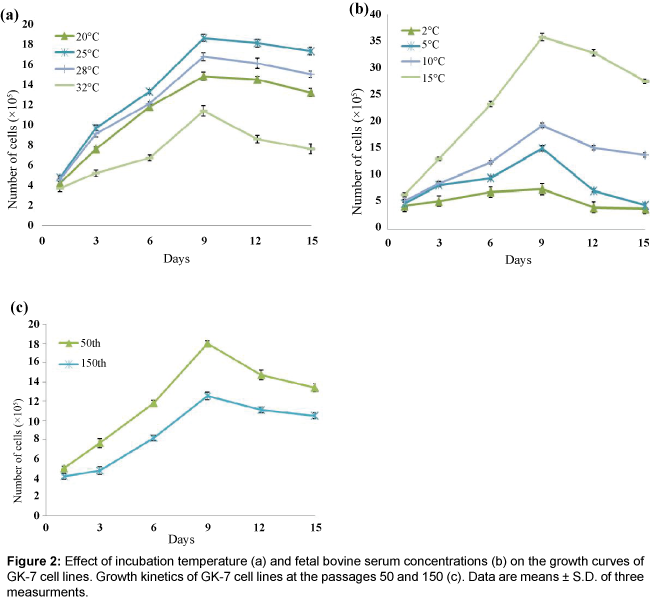
To optimize the conditions for cultivation of the GK-7 cell, the cells at the 50th passage were used to investigate the kinetics of cell growth. The GK-7 cells were able to grow at temperatures between 20�?? and 32�??, and the highest growth occurred at 25�?? (Figure 2a). Growth rates were remarkably reduced at 32�??, indicating that the GK-7 cells were quite sensitive to high temperature. Growth of GK-7 cells at varying levels of FBS concentrations is shown in Figure 2b. Results show that serum is required for the cells to survive and grow and the growth rates of the cell increased with the FBS concentration. With regard the growth rates of GK-7 cell with different passage history, cells of the 50th passage proliferated significantly faster than those of the 150th passage during the observation period of 15 days. The cells of the 50th passage peaked at approximately 1.8 × 106 cells/ml after 9 days of seeding and the cells of the 150th passage gave1.2 × 106 cells/ml at the same day (Figure 2).
Chromosome number analysis
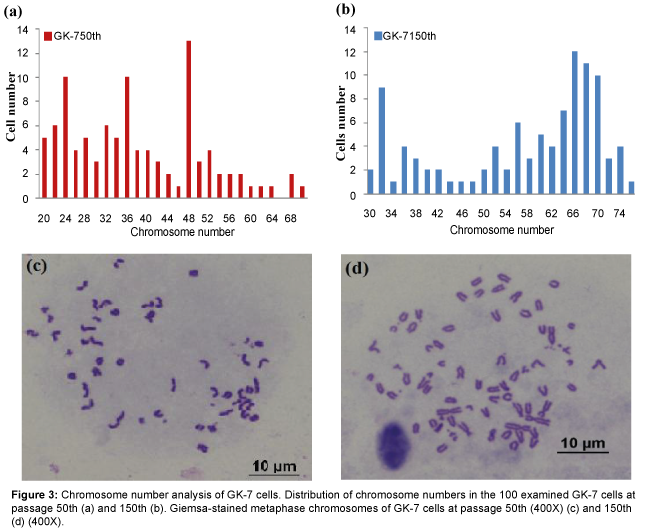
The results of chromosomes counts of 100 metaphase plates at the 50th passage showed diploid numbers ranging from 20 to 68 with a modal peak at 48 chromosomes and the metaphates at the 50th passage with the normal diploid number, additionally displayed the normal karyotype morphology of GK-7 cells consisted of a pair of subtelocentrics (st) and 23 pairs of telocentrics. The chromosome number distribution at the 150th passage based on 100 metaphase plates, displayed 2n values ranging between 30 and 76 with the modal value of 66 chromosomes and had karyotype composed of two pairs st and 31 pairs telocentrics (Figure 3).
Characterization of the GK-7 cell line
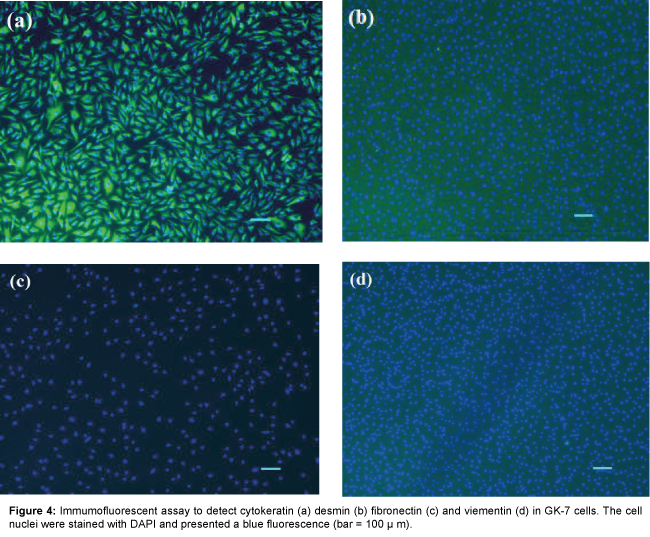
Four antibodies against cytokeratin, vimentin, fibronectin and desmin, respectively, were used for immunostaining to identify the feature of the GK-7 cell. Strong fluorescence signals were observed in the cells reacting with antibody against cytokeratin, an epithelial cell marker (Figure 4). No reactivity was observed in control and with anti-vimentin, anti- fibronectin and anti- desmin antibodies. These results confirmed that the GK-7 cells are epithelial cells.
Identification of cell origin
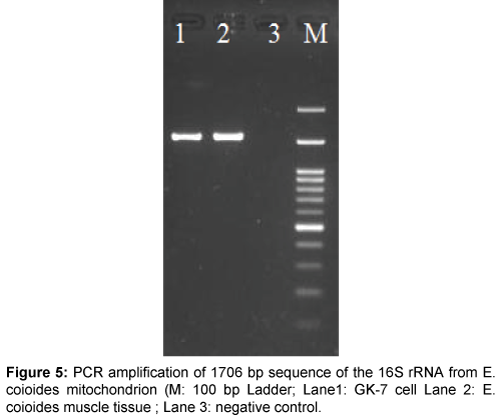
The origin of the GK-7 cell line was examined by the PCR designed for amplifying the conserved region of the 16S rRNA gene. The PCR product of 1706 bp demonstrated a similsr size to that from E. coioides mitochondrial DNA (Figure 5). The nucleotide sequence of the PCR product (GenBank accession number KU510327) was 99.9% similar to the known E. coioides mitochondrial DNA sequences available in GenBank (Accession number KM377093 and EU043376).
Cytopathic effect and virus replication
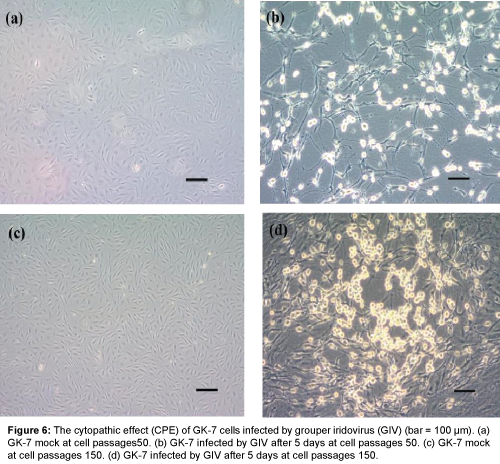
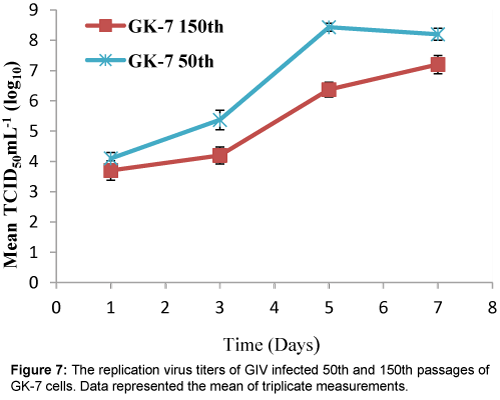
Viral infection was detected in both cell passages at 50th and 150th subcultures of GK-7 cells within 3 days after GIV inoculation. Initially, the specific CPE developed as foci areas of rounded, granular and refractive cells, which spread through the cell sheet over 3-4 days to form a network of degenerated cells and completely disintegrated by 5 days post-inoculation (d.p.i) in the cells of the 50th passage. By contrast, the CPE in the cells of the 150 passage appeared slower than those of the 50 passage (Figure 6). Over 7 days of observation, the virus titers of GK-7 cells of the 50th and 150th passages reached up 108.4 and 107.2 TCID50 mL-1, respectively (Figure 7).
Electron microscopy
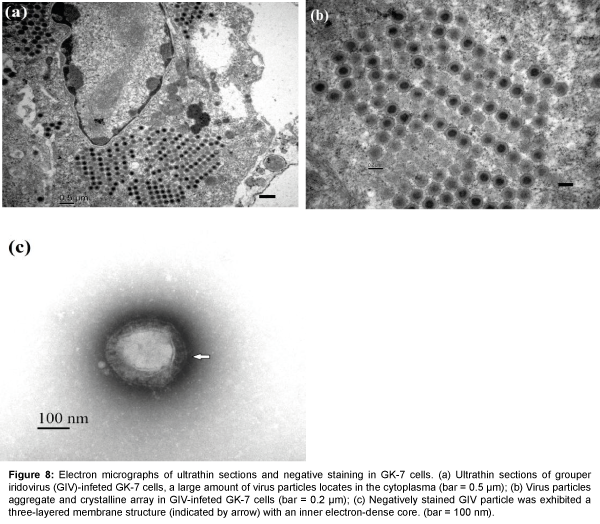
Ultrathin sections of the GIV-infected GK-7 cells at 2 d.p.i. showed a large number of membrane-bound aggregates and crystalline arrays of GIV particles in the cytoplasm (Figure 8). The nuclear membranes remained largely intact, and chromatin margination and cytoplasmic changes were evident in the GK-7 cells (Figure 8a). The virions were hexagonal and crystalline aggregation in infected cells and the mean diameter of the virions were about 200 nm (Figure 8b). Negatively stained enveloped virus showed a typical hexagonal shape, and the envelope structure of virus particle was three-layered with an inner electron-dense core surrounded by a lighter coat (Figure 8c). The particles were larger than those found in ultrathin sections, with a mean diameter about 240 nm.
Figure 8: Electron micrographs of ultrathin sections and negative staining in GK-7 cells. (a) Ultrathin sections of grouper iridovirus (GIV)-infeted GK-7 cells, a large amount of virus particles locates in the cytoplasma (bar = 0.5 μm); (b) Virus particles aggregate and crystalline array in GIV-infeted GK-7 cells (bar = 0.2 μm); (c) Negatively stained GIV particle was exhibited a three-layered membrane structure (indicated by arrow) with an inner electron-dense core. (bar = 100 nm).
Cell transfection
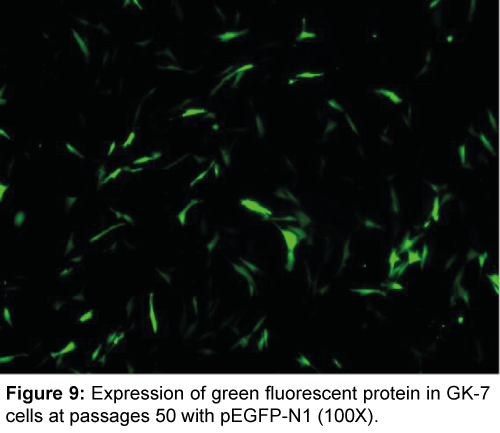
The GK-7 cell line was successfully transfected with pEGFP-N1 vecter by lipofectamine 2000. The green fluorescence signals were observed in GK-7 cell line as early as 24 h post-transfection, and showed clear and strong fluorescence signals after 48 h (Figure 9). The transfection efficiency was around 50%, calculated by microscopy.
Discussion
In this study, we established and characterized a novel cell line from the kidney of E. coioides, and the cell line was designated as GK- 7. Morphologically, the GK-7 cells were composed of spindle-shape epithelial-like cells which was distinct from the GK cells derived from the E. awoara composed of a heterogeneous mixture of fibroblasts and epithelial-like cells [13]. Furthermore, the immunocytochemistry demonstrated strong positivity to cytokeratin which are used as specific markers for epithelial cells. These results indicated that the GK-7 cells were of epithelial origin. Similar results were reported in GBC4 clonal cell line from the brain of E. coioides [23] and EAGL drived from the liver of E.akaara [15].
The GK-7 cells grew efficiently at 20-32�?? and showed the optimum growth at 25�??, which was consistent with the conditions for the other established Epinephelus cell lines [9]. The advantages of cell lines that grow over a wide temperature range is their potential suitability for isolating both warm-water and cold-water fish viruses [14]. Base on the results show that serum is the higher concentration of FBS, the faster growth rate of cells. However, 2% FBS can support maintenance growth and at least required serum proportion for the GK-7 cell line at low cost.
Growth curves between the GK-7 cells of the 50th and 150th were similar. The results of growth curves performed in GK-7 cell line were more stable. In addition, the cells of the 50th passage displayed higher densitiy than that of the cells of the 150th pssages, showing that the GK-7 cell line with shorter passage history could be propagated more rapidly for performing the culture. In addition, our virus replication data showed that the maximum virus yield in the cell of the 50th passage was faster and higher than that in the cells of 150th passage. Maximum virus yield was produced in the GK-7 cells of the 50th and 150th passage at 5 and 7 d.p.i., respectively. The higher production could result from higher cell density present at the time of infection. These results demonstrated the GK-7 cells of the 50th passage was suitable for both research and product manufacture.
In this study, the GK-7 cell line at the 50th and 150th passages had different chromosome numbers. At the 50th passage, the GK-7 cell contained a modal value of 48 chromosomes; in contrast, at the 150th passage, chromosome numbers varied in a range from 30 and 76 with the modal value at 66 chromosomes. The similar chromsome variability is also observed in Epinephelus akaara grouper brain cell line [8]. The modal number of the grouper brain cell line is 48 at the 35th passage and becomes 60 at the 70th passage. A clonal cell line, GBC4, derived from grouper (Epinephelus coioides) brain also demonstrates a shift of chromosome number from 22 to 58 [24]. These results indicated that GK-7 cell line similar to most of the established fish cell line, had undergone some degree of cell transformation.
The cell line used to propagate a virus has to be rapid and efficient propagate the virus in large quantities and should be suitable for a wide variety of virus strains. Our study indicated that the GK-7 cell line was susceptible to GIV and was a statisfactory tool to propagate GIV, as showing that viral titers reached 108.4 TCID50 mL-1. Similar findings have been also reported in the other grouper kidney cell line [13]. These observations indicated that the GK-7 cell line exhibited higher susceptibility to piscine ranaviruses. Enveloped GIV particles were found in the infected cells. Previous ultrastructural studies on ranaviruses reveal that the enveloped virus particles can be seen in the early stages of frog virus 3 (FV3) and SGIV infection [1,17,18]. We first discovered that the GIV also possessed an envelope structure (Figure 8c). In addition, most matured virus particles appeared in the cytoplasm of GIV-infected GK-7 cells (Figure 8a). Similar findings were also reported in a giant grouper (Epinephelus lanceolatus) cell derived from heart tissue [5]. In the cells infected, numerous virus particles were observed in the viral factory and in the cytoplasm near the metamorphic nucleus [5]. The ultrastructure and size of GIV are very similar to those pscine and amphibian iridovirus belonging to genus Ranavirus.
The 16S rRNA has been used to identify the origin of various established cells in previous studies [21]. Our data showed that the sequence of GK-7 16S rRNA matched perfectly with the sequence of orange spotted grouper E. coioides, 16S rRNA [25,26]. In the absence of 16S rRNA data for E.coioides, this is a strong evidence to support that the GK-7 cell line was derived from orange spotted grouper.
The GK-7 cell line we developed allowed foreign genetic materials expressed. Exogenous DNA delivery of a cultured cell is very useful for both basic research and biotechnological applications. In this study, the GK-7 cell line was successfully transfected with pEGFP-N1 plasmid by means of lipofectamine 2000 and transfection efficiency was up to 50% (Figure 9). These results illustrated that this cell line may be able to be genetically manipulated for host-pathogen interactions study.
The present study indicated that the GK-7 cell line is a suitable tools for studying epidemiology and pathology but also can further be applied to develop vaccine to control the GIV-associated diseases.
Acknowledgment
The authors thanks Dr. Fan Lee for his comments on this manuscript. This study was granted by projects 99AS-1.1.5-HI-H2, 100AS-1.1.5-HI-H2, 101AS- 10.2.4-HI-H1, 102AS-10.2.4-HI-H1, 103AS-10.1.3-HI-H1 of the Council of Agriculture, Executive Yuan, Taiwan.
References
- Barunwald J, Nonnenmacher H, Tripier-Darcy F (1985) Ultrastructural and biochemical study of frog virus 3 uptake by BHK-21 cells. J of General Virology 66: 283-293.
- Cheng TC, Lai YS, Lin IY, Wu CP, Chang SL, et al. (2010) Establishment, characterization, virus susceptibility and transfection of cell lines from cobia, Rachycentron canadum (L), brain and fin. J Fish Dis 33: 161-169.
- Chi SC, Hu WW, Lo BJ (1999) Establishment and characterization of a continuous cell line (GF-1) derived from grouper, Epinephelus coioides (Hamilton): a cell line susceptible to grouper nervous necrosis virus (GNNV). Journal of Fish Diseases 22: 173-182.
- Gong J, Huang Y, Huang X, Ouyang Z, Guo M, et al. (2011) Establishment and characterization of a new cell line derived from kidney of grouper, Epinephelus akaara (Temminck & Schlegel), susceptible to Singapore grouper iridovirus (SGIV). J Fish Dis 34: 677-686.
- Guo CY, Huang YH, Wei SN, Ouyang ZL, Yan Y, et al. (2015) Establishment of a new cell line from the heart of giant grouper, Epinephelus lanceolatus (Bloch), and its application in toxicology and virus susceptibility. Journal of Fish Diseases 38: 175-186.
- Huang C, Zhang X, Gin KYH, Qin QW (2004) In situ hybridization of a marine fish virus, Singapore grouper iridovirus with a nucleic acid probe of major capsid protein. J of Virological Methods 117: 123-128.
- Huang SM, Tu C, Tseng CH, Huang CC, Chou CC, et al. (2011) Genetic analysis of fish iridoviruses isolated in Taiwan during 2001–2009. Archives of Virology 156: 1505-1515.
- Huang X, Huang Y, Ouyang Z, Qin Q (2011) Establishment of a cell line from the brain of grouper (Epinephelus akaara) for cytotoxicity testing and virus pathogenesis. Aquaculture 311: 65-73.
- Huang XH, Huang YH, Sun JJ, Han X, Qin Q (2009) Characterization of two grouper Epinephelus akaara cell lines: application to studies of Singapore grouper iridovirus (SGIV) propagation and virus-host interaction. Aquaculture 292: 172-179.
- Ku CC , Teng YC, Wang CS, Lu CH (2009) Establishment and characterization of three cell lines derived from the rockfish grouper Epinephelus quoyanus: Use for transgenic studies and cytotoxicity testing. Aquaculture 294: 147-151.
- Lai YS, John JA, Lin CH, Guo IC, Chen SC, et al. (2003) Establishment of cell lines from a tropical grouper, Epinephelus awoara
- Lai YS, Murali S, Chiu HC, Ju HY, Lin YS, et al. (2001) Propagation of yellow grouper nervous necrosis virus (YGNNV) in a new nodavirus-susceptible cell line from yellow grouper, Epinephelus awoara (Temminck & Schlegel), brain tissue. Journal of Fish Diseases 24:299-309.
- Lai YS, Murali S, Ju HY, Wu MF, Guo IC, et al. (2000) Two iridovirus-susceptible cell lines established from kidney and liver of grouper, Epinephelus awoara (Temminck & Schlegel), and partial characterization of grouper iridovirus. J of Fish Diseases 23: 379-38.
- Nicholson BL, Danner DJ, Wu JL (1987) Three New Continuous Cell Lines from Marine Fishes of Asia. In Vitro Cellular & Developmental Biology 23: 199-204.
- Ou-Yang ZL, Huang XH, Huang EY, Huang YH, Gong J, et al. (2010) Establishment and characterization of a new marine fish cell line derived from red-spotted grouper Epinephelus akaara. Journal of fish biology 77: 1083-1095.
- Parameswaran V, Shukla R, Bhonde RR, Hameed AS (2007) Development and characterization of two new cell lines from milkfish (Chanos chanos) and grouper (Epinephelus coioides) for virus isolation. Marine Biotechnology 9: 281-291.
- Qin QW, Chang SF, Ngoh-Lim GH, Gibson-Kueh S, Shi C, et al. (2003) Characterization of a novel ranavirus isolated from grouper Epinephelus tauvina. Diseases of Aquatic Organisms 53: 1-9.
- Qin QW, Lam TJ, Sin YM, Shen H, Chang SF, et al. (2001) Electron microscopic observations of a marine fish iridovirus isolated from brown-spotted grouper, Epinephelus tauvina. Journal of Virological Methods 98: 17-24.
- Qin QW, Wu TH, Jia TL, Hegde A, Zhang RQ (2006) Development and characterization of a new tropical marine fish cell line from grouper, Epinephelus coioides susceptible to iridovirus and nodavirus. J ofVirological Methods 131: 58-64
- Reed LJ, Muench H (1938) A Simple Method of Estimating Fifty Percent Endpoints. American J of Epidemiology 27: 493-497.
- Wang G, Lapatra S, Zeng L, Zhao Z, Lu Y (2003) Establishment, growth, cryopreservation and species of origin identification of three cell lines from white sturgeon, Acipenser transmontanus. Methods in Cell Science 25: 211-220.
- Wei Y, Fan T, Jiang G, Sun A, Xu X, et al. (2009) Establishment of a novel fin cell line from Brown-marbled grouper, Epinephelus fuscoguttatus (Forsskål), and evaluation of its viral susceptibility. Aquaculture Research 40: 1523-1531.
- Wen CM, Huang JY, Ciou JH, Kao YL, Cheng YH (2009) Immunochemical and molecular characterization of GBC4 as a tanycyte-like cell line derived from grouper brain. Comparative biochemistry and physiology 153: 191-201.
- Wen CM, Lee CW, Wang CS, Cheng YH, Huang HY (2008) Development of two cell lines from Epinephelus coioides brain tissue for characterization of betanodavirus and megalocytivirus infectivity and propagation. Aquaculture 278: 14-21.
- Zhou GZ, Li ZQ, Yuan XP, Zhang QY (2007) Establishment, characterization, and virus susceptibility of a new marine cell line from red spotted grouper (Epinephelus akaara). Marine Biotechnology 9: 370-376.
- Zhuang X, Ding S, Wang JUN, Wang Y, Su Y (2009) A set of 16 consensus primer pairs amplifying the complete mitochondrial genomes of orange-spotted grouper (Epinephelus coioides) and Hong Kong grouper (Epinephelus akaara). Molecular Ecology Resources 9: 1551-1553.
Relevant Topics
- Algal Blooms
- Blue Carbon Sequestration
- Brackish Water
- Catfish
- Coral Bleaching
- Coral Reefs
- Deep Sea Fish
- Deep Sea Mining
- Ichthyoplankton
- Mangrove Ecosystem
- Marine Engineering
- Marine Fisheries
- Marine Mammal Research
- Marine Microbiome Analysis
- Marine Pollution
- Marine Reptiles
- Marine Science
- Ocean Currents
- Photoendosymbiosis
- Reef Biology
- Sea Food
- Sea Grass
- Sea Transportation
- Seaweed
Recommended Journals
Article Tools
Article Usage
- Total views: 12592
- [From(publication date):
June-2016 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 11644
- PDF downloads : 948