ERK5 Silencing Inhibits Invasion of Human Osteosarcoma Cell via Modulating the Slug/MMP-9 Pathway
Received: 13-Jun-2014 / Accepted Date: 17-Jul-2014 / Published Date: 19-Jul-2014 DOI: 10.4172/2161-0681.1000182
Abstract
Background and aim: ERK5 is over expressed in a many of human cancers and this overexpression has been associated with metastasis and invasion. Furthermore, ERK5 siliencing inhibits aggressive phenotypes of cancer cells. However, mechanisms by which ERK5 regulates tumour progression or metastasis have not been elucidated. In this study, using human osteosarcoma cell lines U2OS as a model, we explored the involvement of ERK5 siliencing on invasiveness of U2OS cells.
Materials and methods: ERK5 siRNA targeting ERK5 was stably transfected into the human osteosarcoma cell lines U2OS. ERK5 knocked-down U2OS cells was then transfected with slug cDNA or MMP-9 cDNA plasmid to reexpress Slug or MMP-9. Cell proliferation was detected by MTT assay. Cell invasion and metastasis was detected by Matrigel invasion and wound healing assay. An orthotopic nude mouse model of U2OS was applied for in vivo lung metastasis experiments. ERK5, Slug, MMP-9 and E-cadherin were analyzed by real-time PCR, and Western blotting.
Results: ERK5 silencing by siRNA in U2OS cells decreased Slug and MMP-9 expression. Compared with the vector-transfected cells, ERK5 knocked-down cells showed reduced migration and invasion in vitro, as well as decreased metastatic potential in experimental metastasis. Re-expression of Slug or MMP-9 in ERK5 knocked-down cells restored the invasive phenotypes. We also discovered that Re-expression of Slug in ERK5 knocked-down cells restored the MMP-9 expression, and re-expression of MMP-9 in ERK5 knocked-down cells did not affect slug and ERK5 expression. CONCLUSIONS: Our data suggest that ERK5 knockdown inhibits aggressive behaviour of human U2OS cells through modulating Slug signalling and MMP-9 expression.
Keywords: Osteosarcoma; Invasion; ERK5; Slug; MMP-9
314910Introduction
The MEK5/ERK5 pathway is the least well studied MAPK signalling module. It has been proposed to play a role in the pathology of cancer [1]. Similar to other MAPKs, ERK5 is present in a wide variety of cell types and is believed to be ubiquitously expressed. Presumably, it serves to regulate diverse functions depending on the cellular context and circumstances [2]. ERK5 has been implicated in the survival response of PC12 cells to oxidative stress [3]. Further work demonstrated that ERK5 contributes to the survival response in neuronal dorsal root ganglia cells through a unique retrograde signalling system mediate by nerve growth factor (NGR) [4,5].
Work has been done to examine vascular integrity and endothelial failures in animal studies. ERK5 deletion was embryonically lethal in genetic knock-out mice and ERK5 deletion in adult mice lead to lethality within 2–4 weeks. Physiological analysis of the adult mice demonstrated abnormally leaky blood vessels. Histologically these mice demonstrated multi organ haemorrhage and architectural irregularities in the endothelial lining of their blood vessels. This evidence suggests that ERK5 is critical for endothelial function and preserves the integrity of blood vessels [6]. Recent study found that ERK5 also plays a role in controlling cytoskeleton organization and motility of keratinocytes during cutaneous wound healing [7]. However, mechanisms by which ERK5 promotes tumour progression or metastasis have not been elucidated.
Epithelial to mesenchymal transition (EMT), which has been recognized for several decades as critical for embryogenesis [8], has recently been shown to also be relevant to cancer progression. During EMT of in situ cancer cells, expression of proteins that promote cell–cell contact such as E-cadherin can be lost, and mesenchymal markers such as vimentin, fibronectin, Ncadherin and the metalloproteinases MMP-2 and MMP-9 can be acquired, resulting in enhanced ability for cell migration and invasion [9]. Identified EMT-driving transcription factors include the zinc-finger-containing Snail family members (e.g. Snail and Slug), ZEB family members (e.g. ZEB1, SIP1), and basic helix–loop–helix (bHLH) factors (e.g. E2A, Id2, Id3, Twist); these factors serve as transcriptional repressor of the E-cadherin gene [9].
A study investigated the role of ERK5 in reepithelialisation during cutaneous wound healing of human keratinocytes. The cells were treated with EGF to stimulate the EGF receptor. Treated cells showed increased levels of phosphorylated ERK5 which coincided with increased levels of Slug mRNA [7]; An ERK5 knockdown was also used in HaCaT cells, an immortalized human keratinocyte cell line. The treated HaCaT cells demonstrated decreased motility response and reduced Slug mRNA expression. The knockdown cells demonstrated a more compact morphology, disruption in desmosome organization and an altered ability to aggregate. This work suggests that ERK5 plays a role in controlling cytoskeleton organization and motility of keratinocytes by regulation of Slug during cutaneous wound healing [7].
It has recently found Slug promotes migration and invasion of PANC-1 cells, which may correlate with the reorganization of MMP-9 and remodelling of the F-actin cytoskeleton, but not with E-cadherin expression [10]. Kim et al. has found ERK5 regulates the invasion of OS cells by inducing MMP-9 expression [11].
Osteosarcoma (OS) is a type of aggressive bone cancer of mesenchymal origin generally found in youths aged between 10 and 25 years. Any bone of the human body can be affected by this neoplasia and the general survival at five years is approximately 65 to 75%. The main causes of death are pulmonary metastases diagnosed by computed tomography (CT) in 35 to 45% of the patients [12]. In this study, aiming at further understanding the mechanisms underlying the progression and metastasis of OS, we employed an OS cell line and explored the role of ERK5 in modulating the invasive phenotype of cancer cells. We provide evidence linking ERK5 to the regulation the invasion and metastasis through the Slug /MMP-9 pathway in human OS cells.
Materials and Methods
Cell culture
Human osteosarcoma cell lines U2OS cell lines was purchased from the American Type Culture Collection and maintained in McCoy's 5A medium (Invitrogen). The cells were incubated at 37°C in humidified air containing 5% v/v CO2.
Plasmid constructs
MMP-9 cDNA was from Sino Biological Inc. BamH I and EcoR I enzyme were used to digest the MMP-9 cDNA gene and pcDNA3.1 plasmid. MMP-9 cDNA gene was inserted (Invitrogen) into plasmids pcDNA3.1 to construct pcDNA3.1- MMP-9 cDNA. The structures and fidelity of the resulting constructs were confirmed by restriction mapping and sequencing. Plasmids were purified using the Plasmid Midi kit (Qiagen). At least two independent plasmid preparations of each construct were used in reporter assays. The control pcDNA3.1 plasmid was construced as the method of above.
Cell transfection
The ERK5 siRNA was purchased from Dharmacon Inc. (Lafayette, CO). A non-targeting siRNA (control siRNA; Dharmacon Inc.) was used as a negative control. To generate ERK5-silenced stable clones (U2OS-ERK5 siRNA 1 and 2), U2OS cells were transfected with 100 nM/L of control RNA or ERK5 siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer instructions. The cells were harvested 48 hours after siRNA transfection. siRNA knockdown efficiency was measured by real time RT-PCR and Western blot.
For clones of U2OS-ERK5 siRNA cells expressing MMP-9 or Slug, U2OS-ERK5 siRNA 1/2 cells were transfected with pcDNA3.1- MMP-9 cDNA or pcDNA3.1-SLUG cDNA (kindly donated by Dr. Zhang, the Affiliated Hospital of QingDao University), and pcDNA3.1 as a control using Lipofectamine 2000 (Invitrogen). For selection of stably transfected cell populations, G418 was added to the culture medium 48 h after transfection at a concentration of 400 μg ml-1 for the above transfected U2OS cells. The cells were selected with 400 μg/mL G418 for 14 d.
Real-Time RT-PCR
Real-time RT-PCR (QPCR) was performed using the LightCycler as the manufacture’s instruction. For a final reaction volume of 20 μl, the following reagents were added: 10 μl SYBR Advantage qPCR Premix (Clontech, Mountain View, CA), 1 μL of each forward and reverse 10 μM primers (Table 1), 7 μL H2O, and 1 μL cDNA template. Messenger RNA (mRNA) levels were quantified using the standard curve method. Standard curves were constructed by using serially diluted standard template. The data were normalized to 18S ribosomal RNA or ß-actin RNA to account for differences in reverse transcription efficiencies and the amount of template in the reaction mixtures.
| Gene | Sequence 5’-3’ |
|---|---|
| ERK5 | CTGGCTGTCCAGATGTGAA ATGGCACCATCTTTCTTTGG |
| Slug | GCGAACTGGACACACACACAGTTAT CCCCAGTGTGAGTTCTAATGTGTCC |
| E-cadherin | GGAAGTCAGTTCAGACTCCAGCC AGGCCTTTTGACTGTAATCACACC |
| MMP-9 | CCTTCTACGGCCACTACT GCACTGCAGGATGTCATA |
| β-actin | CCTCTATGCCAACACAGTGC CATCGTACTCCTGCTTGCTG |
| 18S | GTAACCCGTTGAACCCCATT CCATCCAACGGTAGTAGCG |
Table 1: Oligonucleotide Sequences for Real Time RT-PCR
Western blot analysis
Cells were washed twice with PBS and lysed in the RIPA buffer (Upstate) at 4°C for 20 min. Lysates were cleared at 12,000 g for 12 min at 4°C and supernatants were subjected to western analysis. Equal-amount (40 μg) cell lysates were separated by SDS-PAGE (10% gel), transferred to polyvinylidene membranes (Millipore), and probed with antibodies against proteins of interest, including Phospho-ERK5 antibody (pERK5) and ERK5 antibodyCell Signaling Techn, E-cadherin (Biosciences), MMP-9 (Santa Cruz Biotechnology) and ß-actin (Abcam). Blots were then incubated with appropriate horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Inc.). Bound antibodies were visualized by enhanced chemiluminescence.
Cell proliferation assay
Cells were seeded at 5×103 per well on 96-well plates in growth medium supplemented with 10% serum, and were cultured in a humidified chamber at 37°C for up to 3 days. Viable cells were identified using the MTT assay as the manufacture’s instruction. The contents of the plates were mixed for 5 min, and the absorbance was read at 540 nm using a plate reader.
Matrigel invasion assay
Cell invasion assays were performed using Transwell membrane filter inserts with 8-mm pore size (Corning Costar, Cambridge, MA). Cells were trypsinized, washed and resuspended in RPMI-1640+10% FBS. Samples of 50,000 cells were placed in the upper chamber of each Transwell device (Nucleopore) with 8-μm Matrigel-coated polycarbonate membrane filter insert in 24-well plates, and the same medium was placed in the lower chamber. After 24 hours of incubation, non-invading cells were removed by wiping the upper surface of the filter with a cotton swap; the remaining cells were fixed in 100% methanol for 20 min and stained with GIEMSA (Sigma). The degree of invasion was quantified by counting the cells on the underside of filters under a microscope. Experiments were repeated at least three times in duplicates for each cell line.
Wound healing assay
Cells were seeded at 1×106 cells per well in 6-well plates and cultured for approximately 24 h to be nearly confluent. A scratch “wound”, i.e. a cell-free line of 20-cell diameters in width, was made using a p200 pipette tip and the medium was changed to remove detached cells. Phase contrast micrographs were taken immediately and 24 h after making the wound. Numbers of cells which have migrated were counted comparing the micrographs. For each sample, cells from three representative fields were counted.
In vivo experimental lung metastasis
For experimental lung metastasis assay, 1×106 cells resuspended in 100 μl PBS buffer were injected into the tail vein of 7- to 10-week-old male non-obese diabetic severe combined immunodeficient (NODSCID) mice. Mice were sacrificed 8 weeks after injection. Lungs were removed and fixed in 10% formalin. Paraffin-embedded lung tissues were sectioned and stained with haematoxylin and eosin for histological examination. The extent of lung metastasis was scored by counting total tumour nodules in serial 5-μm sections
Statistical analysis
Data are shown as the mean ± S.D. where possible and statistical analysis was obtained using SPSS version 11.0, computer software (SPSS Inc., Chicago, IL, USA). A P-value of <0.05 was taken to indicate statistical significance. Statistical significance of difference between groups was tested using Student’s t-test or if there were more than two groups, using one way analysis of variance (ANOVA) followed by posthoc analysis.
Results
No effect of ERK5 knockdown on cell proliferation
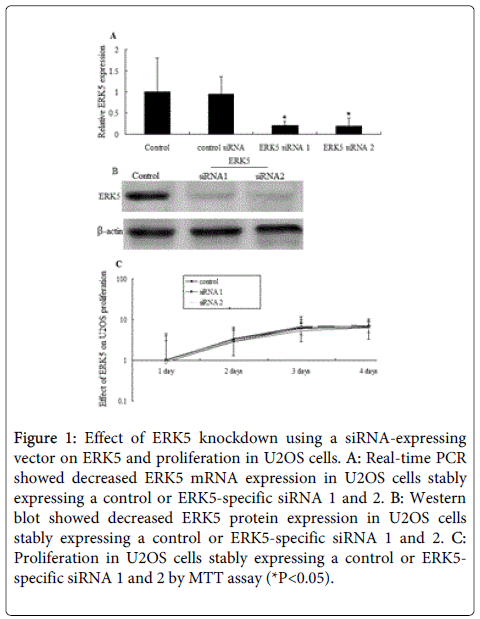
Both qRT-PCR and Western blot analyses revealed higher ERK5 expression levels in U2OS cell. The degree of inhibition of ERK5 mRNA expression induced by siRNA against ERK5 is shown in Figure 1A. SiRNA 1 and siRNA 2 targeting ERK5 inhibited ERK5 mRNA levels in siRNA stably transfected U2OS cell to <10% of control cells, respectively (Figure 1A). Similar results were observed in ERK5 protein levels after siRNA 1 and siRNA 2 stable transfection by western blot assay (Figure 1B).
Figure 1: Effect of ERK5 knockdown using a siRNA-expressing vector on ERK5 and proliferation in U2OS cells. A: Real-time PCR showed decreased ERK5 mRNA expression in U2OS cells stably expressing a control or ERK5-specific siRNA 1 and 2. B: Western blot showed decreased ERK5 protein expression in U2OS cells stably expressing a control or ERK5-specific siRNA 1 and 2. C: Proliferation in U2OS cells stably expressing a control or ERK5- specific siRNA 1 and 2 by MTT assay (*P<0.05).
Inhibition of ERK5 expression in U2OS cells did not suppress cell proliferation compared to the control after 4 days after stable transfection (Figure 2C). The information indicated that ERK5 knockdown did not affect the growth curves of U2OS cells.
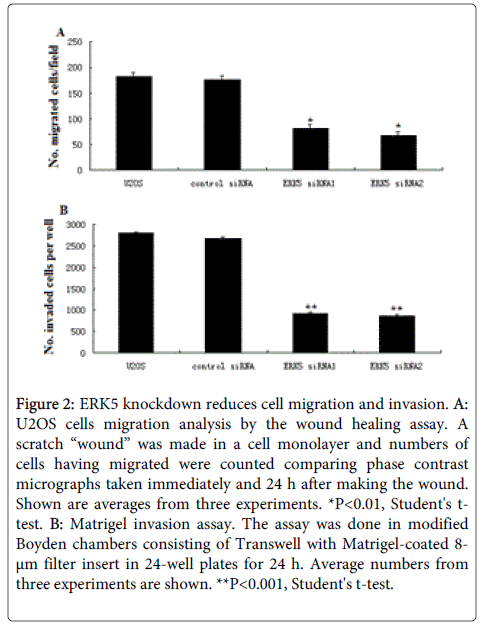
Figure 2: ERK5 knockdown reduces cell migration and invasion. A: U2OS cells migration analysis by the wound healing assay. A scratch “wound” was made in a cell monolayer and numbers of cells having migrated were counted comparing phase contrast micrographs taken immediately and 24 h after making the wound. Shown are averages from three experiments. *P<0.01, Student's ttest. B: Matrigel invasion assay. The assay was done in modified Boyden chambers consisting of Transwell with Matrigel-coated 8- μm filter insert in 24-well plates for 24 h. Average numbers from three experiments are shown. **P<0.001, Student's t-test.
ERK5 knockdown on migration, invasion, and metastasis of U2OS cells
ERK5 knockdown in U2OS did not affect the growth curves of cells .We next examined the effect of ERK5 knockdown on the invasive behaviour of cells. When compared to control cells, ERK5 siRNA cells showed less migration in the wound-healing assay (Figure 2A) and exhibited decreased invasion in the Matrigel assay (Figure 1B). To check if the reduced in vitro migration and invasiveness in ERK5 siRNA clones could affect the in vivo metastatic potential of cells, we performed an experimental metastasis assay. Severe combined immunodeficiency (SCID) mice were injected with ERK5 siRNA 1 and ERK5 siRNA2, vector transfected U2OS cells respectively via the tail veins, and examined 7 weeks after injection for metastatic nodules formed in lungs. While most mice injected with the parental or vector transfected U2OS cells formed pulmonary tumour nodules, none of the mice injected with ERK5 siRNA cells developed metastatic lesions in lungs (Table 2), demonstrating that ERK5 knockdown reduced the metastatic potential of U2OS cells.
| U2OS | Control siRNA | ERK5 siRNA1 | ERK5 siRNA2 | |
|---|---|---|---|---|
| No. of mice with PN/ No. of mice injected | 8/10 | 7/9 (one mouse died) | 0/10* | 0/9* (one mouse died) |
| Average No. of PN per mouse±s.d | 7.1±3.2 | 7.8±3.4 | 0** | 0** |
Table 2: ERK5 knockdown decreases the metastatic potential of U2OS cells. PN: pulmonary nodules. Asterisks indicate statistically significant difference between the Control siRNA and ERK5 siRNA1/2 groups (*P<0.05, one-way ANOVA; **P<0.01, Student's t-test).
ERK5 knockdown suppresses the Slug-dependant MMP-9 expression in the U2OS cells
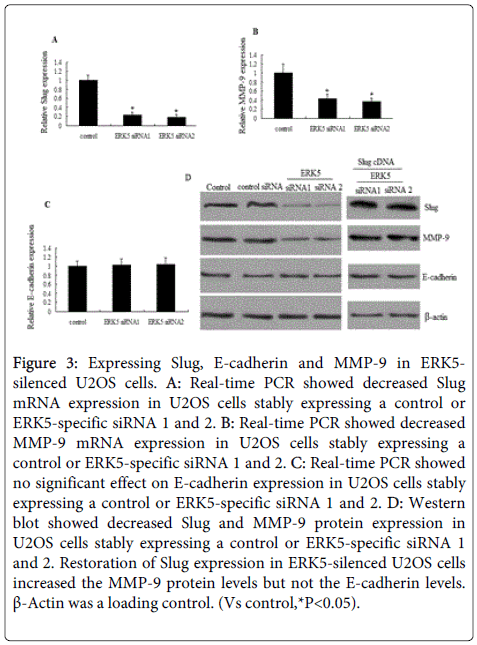
qRT-PCR analysis revealed that ERK5 knockdown inhibits Slug (Figure 3A) and MMP-9 mRNA (Figure 3B) levels, but ERK5 knockdown did not induce E-cadherin upregulation (Figure 3C). Similar results was found by western blot assay (Figure 3D).
Figure 3: Expressing Slug, E-cadherin and MMP-9 in ERK5- silenced U2OS cells. A: Real-time PCR showed decreased Slug mRNA expression in U2OS cells stably expressing a control or ERK5-specific siRNA 1 and 2. B: Real-time PCR showed decreased MMP-9 mRNA expression in U2OS cells stably expressing a control or ERK5-specific siRNA 1 and 2. C: Real-time PCR showed no significant effect on E-cadherin expression in U2OS cells stably expressing a control or ERK5-specific siRNA 1 and 2. D: Western blot showed decreased Slug and MMP-9 protein expression in U2OS cells stably expressing a control or ERK5-specific siRNA 1 and 2. Restoration of Slug expression in ERK5-silenced U2OS cells increased the MMP-9 protein levels but not the E-cadherin levels. β-Actin was a loading control. (Vs control,*P<0.05).
We next investigates whether MMP-9 was dependant on Slug. A Slug-expressing plasmid was transfected into ERK5 siRNA cells. The results showed MMP-9 mRNA and proterin (Figure 3D) expression was significantly increased. But slug re-expression did not induce E-cadherin downregulation. Vector transfection did not affect the MMP-9 levels ( data not shown).
We then investigates whether MMP-9 re-expression could affect Slug and ERK5 levels. A MMP-9-expressing plasmid was transfected into ERK5 siRNA cells. The results showed MMP-9 re-expression did not affect the Slug, E-cadherin and ERK5 levels (data not shown). We therefore suggested that ERK5 knockdown suppresses the Slug-dependant MMP-9 expression in the U2OS cells.
Re-expression of Slug or MMP-9 in ERK5-silenced U2OS restores invasive phenotypes
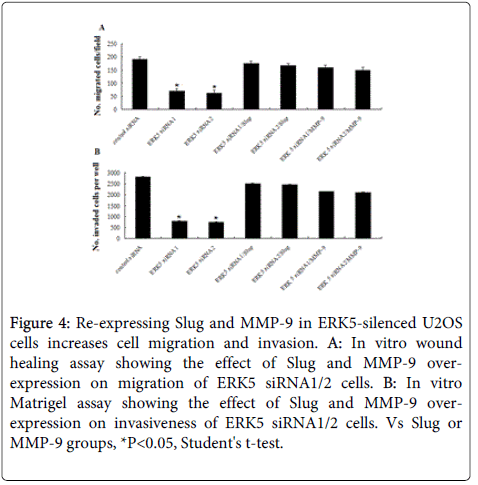
It has shown above that ERK5 siRNA cells showed decreased migration and invasion ability in U2OS cells. At the same time, Slug and MMP-9 expression at both mRNA and protein levels was significantly inhibited in the U2OS cells stably expressing a control or ERK5-specific siRNA 1 and 2 (Figure 3). Compared to control cells, ERK5 siRNA 1/2 cells with reconstituted Slug or MMP-9 expression displayed more migration (Figure 4A) and increased invasion (Figure 4B). The results suggest that ERK5 expression associates with aggressive phenotypes of U2OS cells by regulation of Slug and MMP-9.
Figure 4: Re-expressing Slug and MMP-9 in ERK5-silenced U2OS cells increases cell migration and invasion. A: In vitro wound healing assay showing the effect of Slug and MMP-9 overexpression on migration of ERK5 siRNA1/2 cells. B: In vitro Matrigel assay showing the effect of Slug and MMP-9 overexpression on invasiveness of ERK5 siRNA1/2 cells. Vs Slug or MMP-9 groups, *P<0.05, Student's t-test.
Discussion
In the present study, we observed that the ERK5 knockdown inhibits the in vitro invasiveness and in vivo lung metastasis. In vitro, ERK5 knockdown inhibits the invasiveness through the transcriptional down-regulation of Slug and MMP-9 in human U2OS cells.
The Snail family, which includs Slug, Snail, Snail-like, Scratch1, and Scratch2, has been shown to participate in mesoderm formation, neural crest cell formation and migration, cell differentiation, cell adhesion, cell invasion, cell cycle regulation, and antiapoptosis [13-18]. Slug is expressed in several carcinomas with increased invasion through regulation of E-cadherin [19]. Valerie et al. has found that ERK5 regulats the cytoskeleton by slug signals, and activated ERK5 was the most potent activator of a slug promoter-driven reporter gene. We therefore suggested that ERK5 might be a upstream gene of Slug. In our study, we found that ERK5 and slug was overexpressed in the U2OS cells at gene and protein levels. Knockdown of ERK5 inhibits invasion and metastasis of U2OS cells, followed by decreased slug expression. However, when a slug-expressing plasmid was transfected into the ERK5 siRNA cells, the invasive phenotypes of ERK5 siRNA transfected cells was restored.
Slug is a E-cadherin repressor, which has recently been demonstrated to be important for cancer cells to down-regulate epithelial markers and up-regulate mesenchymal markers in order to become motile and invasive. Birgit et al. has found slug did not repress E-cadherin, neither at the RNA nor at the protein level [20]. Shuji et al. has found using siRNA for Slug demonstrated no effects on cancer cell migration or invasion in vitro [21]. In the present study, we found ERK5 knockdown or slug re-expression did not induce E-cadherin expression. E-cadherin appears to have insignificant role in this process.
MMPs play an important role in degradation of extracellular matrix, which is an essential step in the cascade of metastasis [22]. In the present study, the western blot and QRT-PCR all showed that ERK5 siRNA transfected cells had decreased slug and MMP-9 RNA and protein production, and Slug-transfected cells had increased MMP-9 RNA and protein production, which is a major component of basement membrane. We have also provided evidence suggesting that ERK5 signals through the Slug cascade to regulate MMP-9 expression. However, it remains to be determined how ERK5 links to Slug activation. Relatively little is known about the upstream signaling events that regulate MMP-9 function in cancer cells. More investigation is needed to delineate the signalling mechanism underlying the Slug activation by ERK5.
Conclusion
We conclude that ERK5 siliencing inhibits invasiveness of human OS cells. The mechanism underlying the ERK5-mediated invasiveness involves the activation of Slug and MMP-9 regulation. Further characterization of this ERK5-Slug-MMP-9 signalling pathway to discover its molecular components or regulators should bring about additional insights concerning the molecular action of ERK5 and the complex regulation of the invasion during cancer progression.
References
- Lochhead PA, Gilley R, Cook SJ (2012) ERK5 and its role in tumour development. BiochemSoc Trans 40: 251-256.
- Buschbeck M, Ullrich A (2005) The unique C-terminal tail of the mitogen-activated protein kinase ERK5 regulates its activation and nuclear shuttling. J BiolChem 280: 2659-2667.
- Suzaki Y, Yoshizumi M, Kagami S, Koyama AH, Taketani Y, et al. (2002) Hydrogen peroxide stimulates c-Src-mediated big mitogen-activated protein kinase 1(BMK1) and the MEF2C signaling pathway in PC12 cells: potential role in cell survival following oxidative insults. J BiolChem 277: 9614.
- Watson FL, Heerssen HM, Bhattacharyya A, Klesse L, Lin MZ, et al. (2001) Neurotrophins use the Erk5 pathway to mediate a retrograde survival response. Nat Neurosci 4: 981-988.
- Wang X, Tournier C (2006) Regulation of cellular functions by the ERK5 signalling pathway. Cell Signal 18: 753-760.
- Hayashi M, Kim SW, Imanaka-Yoshida K, Yoshida T, Abel ED, et al. (2004) Targeted deletion of BMK1/ERK5 in adult mice perturbs vascular integrity and leads to endothelial failure. J Clin Invest 113: 1138-1148.
- Arnoux V, Nassour M, L'Helgoualc'h A, Hipskind RA, Savagner P (2008) Erk5 controls Slug expression and keratinocyte activation during wound healing. MolBiol Cell 19: 4738-4749.
- Korsching E, Packeisen J, Liedtke C, Hungermann D, Wülfing P, et al. (2005) The origin of vimentin expression in invasive breast cancer: epithelial-mesenchymal transition, myoepithelialhistogenesis or histogenesis from progenitor cells with bilinear differentiation potential? J Pathol 206: 451-457.
- Kang Y, Massagué J (2004) Epithelial-mesenchymal transitions: twist in development and metastasis. Cell 118: 277-279.
- Zhang K, Chen D, Jiao X, Zhang S, Liu X, et al. (2011) Slug enhances invasion ability of pancreatic cancer cells through upregulation of matrix metalloproteinase-9 and actin cytoskeleton remodeling. Lab Invest 91: 426-438.
- Kim SM, Lee H, Park YS, Lee Y, Seo SW (2012) ERK5 regulates invasiveness of osteosarcoma by inducing MMP-9. J Orthop Res 30: 1040-1044.
- Unni KK (2010) Osteosarcoma. In: Unni KK, editor. Dahlin´s bone tumors (6th Edn): Lippincott Williams & Wilkins, Philadelphia: 122-157.
- Nieto MA (2002) The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol 3: 155-166.
- Savagner P, Karavanova I, Perantoni A, Thiery JP, Yamada KM (1998) Slug mRNA is expressed by specific mesodermal derivatives during rodent organogenesis. DevDyn 213: 182-187.
- CarlTF, Dufton C, HankenJ, KlymkowskyMW (1999) Inhibitionof neural crest migration in Xenopus using antisenseSlug RNA. DevBiol 213: 101-115.
- Catalano A, Rodilossi S, Rippo MR, Caprari P,Procopio A (2004) Induction of stem cell factor/c-Kit/Slugsignal transduction in multidrug-resistant malignantmesothelioma cells. J BiolChem 279: 46706-46714.
- Inoue A, Seidel MG, WuW (2002) Slug, a highly conservedzinc finger transcriptional repressor, protectshematopoietic progenitor cells from radiation-inducedapoptosis in vivo. Cancer Cell 2: 279-288.
- Kajita M, McClinic KN, Wade PA (2004) Aberrant expressionof the transcription factors Snail and Slug alters the response to genotoxic stress. Mol Cell Biol 24: 7559-7566.
- Alves CC, Carneiro F, Hoefler H, Becker KF (2009) Role of the epithelialmesenchymaltransition regulator Slug in primary human cancers.Front Biosci 14: 3035-3050.
- Birgit H, Marco A, Sonja D, Sarah B, Heinz-J B, et al. (2007) Epithelial to Mesenchymal Transition: Expression of the Regulators Snail, Slug, and Twist in Pancreatic Cancer. Clinical Cancer Research 13: 4769.
- Shuji M, Ken-Ichi K, Mototsugu O, Masaru I, Takeo K, et al. (2011) Expression of Snail and Slug in renal cell carcinoma:E-cadherin repressor Snail is associated with cancerinvasion and prognosis. Laboratory Investigation 91: 1443-1458.
- Egeblad M, Werb Z (2002) New functions for the matrixmetalloproteinases in cancer progression. Nat RevCancer 2: 161-174.
Citation: Yue B, Ren QX, Su T, Wang L, Zhang L, et al. (2014) ERK5 Silencing Inhibits Invasion of Human Osteosarcoma Cell via Modulating the Slug/MMP-9 Pathway. J Clin Exp Pathol 4:182. DOI: 10.4172/2161-0681.1000182
Copyright: © 2014 Yue B, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 14798
- [From(publication date): 9-2014 - Apr 04, 2025]
- Breakdown by view type
- HTML page views: 10187
- PDF downloads: 4611