Research Article Open Access
Enzymatic Potential of Mucor inaequisporus for Naringin Biotransformation, Accessed by Fractional Factorial Design and Mass Spectrometry Analysis
Enzo Monte Canedo, Taicia Pacheco Fill*, Edenir Rodrigues Pereira-Filho and Edson Rodrigues-FilhoChemistry Department, Universidade Federal of São Carlos, São Carlos, Brazil
- *Corresponding Author:
- Taicia Pacheco Fill
Chemistry Department, Universidade Federal de São Carlos CP 676
13.565-905, São Carlos, SP, Brazil
Tel: +55-16-3351-8053
Fax: +55-16-3351-8350
E-mail: taicia@gmail.com
Received date: October 4, 2014; Accepted date: October 27, 2014; Published date: October 31, 2014
Citation: Canedo EM, Fill TP, Pereira-Filho ER, Rodrigues-Filho E (2014) Enzymatic Potential of Mucor inaequisporus for Naringin Biotransformation, Accessed by Fractional Factorial Design and Mass Spectrometry Analysis. J Anal Bioanal Techniques S6:006. doi: 10.4172/2155-9872.S6-006
Copyright: © 2014 Canedo EM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
Whole cells of Mucor inaequisporus were used for biotransformation of the flavonoid naringin, as a green method for structural diversification of this class of natural product. The metabolism of the fungus was challenged against naringin under several culturing conditions, using statistical optimization. Eight parameters were evaluated comprising type and concentration of carbon source, substrate concentration, substrate addition time, shaking, luminosity, temperature and extraction time. Sixteen different culture conditions were tested in this screening. After LC-MS analysis, several biotransformation products were identified and naringenin was identified as the major product. Eleven biotransformation products were detected in Ft-HRMS analyses and identified as naringenin as major compound, 4’-methoxynaringenin, 4’-methoxynaringin, rhoifolin, apigenin, methoxyapigenin, acetylated naringin, and also two biflavonoids and two triflavonoids. Ft-HRMSn showed to be a powerful technique for structural elucidation of flavonoid biotransformation products. The fungus was capable to perform deglycosylation, reduction and O-methylation reactions at this substrate indicating good enzymatic potential, since these reactions weren’t observed before for this fungus-genus.
Keywords
Biotransformation; Naringin; Statistical optimization; Mucor inaequisporus; Mass spectrometry; Flavonoid fragmentation; Ft-HRMS
Introduction
Flavonoids are an important and widespread group of plant natural products that possess many biological activities. These compounds are part of the wide range of substances called “polyphenols”, which are widely known mainly by their antioxidant properties, and are present in human dietary sources showing great health benefits [1]. Therefore, researchers are constantly looking for new sources of flavonoids, new structures, and strategies for their interconversions. Flavonoid biotransformation is an important, usually “green”, method for structural modification that may lead to high chemical diversity, which could improve bioavailability and biological properties. In some cases, the microorganisms can mimic mammalian and plant metabolism [2,3].
In recent studies, the biotransformation of naringin, which is a flavanone glycoside present in citrus fruits and grapefruit, and is responsible for the bitterness of citrus juices [4], was performed by several microorganisms using intact cells and isolated enzymes in order to improve some chemical and biological properties. Bacillus stearothermophilus maltogenic amylase, as an example, transglycosylated naringin to maltosylnaringin which is 250 times more soluble in water and 10 times less bitter than naringin [5], whereas a Candida antarctica lipase enhanced its liposolubility by sugar a4cylation [6]. Although several researchers are interested in naringin to naringenin bioconversion using naringinase [7-9] and to prunin [10,11], some studies showed great functionalization of naringin using microbial biotransformation. For instance, naringin metabolism by Penicillium charlesii produced two unknown compounds besides naringenin and prunin [12], whilst Trichoderma harzianum improved naringin antioxidative activity performing a B-ring mono- and dihydroxylation [13]. In other report, Aspergillus saitoi produced carthamidin and isocarthamidin, products of a ring hydroxylation at positions 6 and 8, respectively [14]. Also a C ring opening by Butyrivibrio sp. produced phloroglucinol and p-hydroxyphenylpropionic acid in addition to naringenin and neohesperidose when using intact cells for this microbial transformation [15].
Biotransformation reactions using factorial design approach are frequently used to optimize reactional parameters in some biochemical processes performed by microorganisms. The advantage is to test different conditions in reduced number of experiments, improving the process workout and observing synergic or antagonic effects among variables [16-19]. Our objectives are to study via statistical optimization a way to enhance naringin biotransformation by Mucor inaequisporus and show the capability of mass spectrometry to detect and identify biotransformation products. This is the first report about microbial transformation of naringin by a Mucor species. We found that this fungus was capable to perform deglycosylation, dehydrogenation and O-methylation in naringin, producing eleven biotransformation products.
Materials and Methods
General experimental procedures
The fungus Mucor inaequisporus was isolated from Syzygium cumini (L.) Skeels fruits and identified by sequencing two Internal Transcribed Space (ITS) rDNA regions [20]. The strain has been maintained on malt-agar slants with repeated subculturing. HPLCDiode array detector analyses were performed in a SHIMADZU HPLC equipped with a PDA detector (Tokyo, Japan). HPLC-Electrospray ionization/High resolution mass spectrometry analyses were carried out on a LTQ-Orbitrap Thermo Fisher Scientific mass spectrometry system (Bremen, Germany). Naringin and naringenin were purchased from Aldrich. Fungus growth media were purchased from commercial suppliers and used as provided. All solvents were distilled before use.
Preparation of cultivation medium and culture conditions
The fungus was cultivated in a rich media composed of 50.0 g.L-1 soluble starch or dextrose, 3.0 g.L-1 sodium nitrate, 1.0 g.L-1 potassium phosphate monobasic, 0.5 g.L-1 magnesium sulphate heptahydrate, 0.5 g.L-1 iron sulphate heptahydrate, 0.5 g.L-1 potassium chloride and 20.0 g.L-1 yeast extract. The culture conditions are designed by a 28-4 fractional factorial design experiment described in Table 1. The eight factors (variables) analyzed were (1) type and (2) concentration of carbon source, (3) substrate concentration, (4) substrate addition day, (5) shaking, (6) luminosity, (7) temperature and (8) extraction day. These variables were tested in two levels named -1 (lower level) and +1 (higher level) and more details can be observed at Table 1. The spore suspension of M. inaequisporus was used to inoculate into culture medium using 100 μL of 106 CFU.mL-1 spore suspension. Cultures were grown in 125 mL capacity Erlenmeyer flasks in triplicate and the substrate was prepared in dimethyl sulfoxide.
| 1 Carbon | 2 [Carbon] | 3 [Substr] | 4 Substr. add. time | 5 = 123 Shak. | 6 = 134 Lum. | 7 = 234 Temp. | 8 = 1234 Extr. time | |
|---|---|---|---|---|---|---|---|---|
| f1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | 1 |
| f2 | 1 | -1 | -1 | -1 | 1 | 1 | -1 | -1 |
| f3 | -1 | 1 | -1 | -1 | 1 | -1 | 1 | -1 |
| f4 | 1 | 1 | -1 | -1 | -1 | 1 | 1 | 1 |
| f5 | -1 | -1 | 1 | -1 | 1 | 1 | 1 | -1 |
| f6 | 1 | -1 | 1 | -1 | -1 | -1 | 1 | 1 |
| f7 | -1 | 1 | 1 | -1 | -1 | 1 | -1 | 1 |
| f8 | 1 | 1 | 1 | -1 | 1 | -1 | -1 | -1 |
| f9 | -1 | -1 | -1 | 1 | -1 | 1 | 1 | -1 |
| f10 | 1 | -1 | -1 | 1 | 1 | -1 | 1 | 1 |
| f11 | -1 | 1 | -1 | 1 | 1 | 1 | -1 | 1 |
| f12 | 1 | 1 | -1 | 1 | -1 | -1 | -1 | -1 |
| f13 | -1 | -1 | 1 | 1 | 1 | -1 | -1 | 1 |
| f14 | 1 | -1 | 1 | 1 | -1 | 1 | -1 | -1 |
| f15 | -1 | 1 | 1 | 1 | -1 | -1 | 1 | -1 |
| f16 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Levels (-1 and +1) description | ||||||||
| -1 | Soluble starch | 10 g.L-1 | 5 mg | 1 day | Static | Dark | 25°C | 12 days |
| 1 | Dextrose | 50 g.L-1 | 40 mg | 7 days | 200 rpm | Light | 35°C | 20 days |
Table 1: 28-4 fractional factorial design for biotransformation of naringin by M. inaequisporus with factors and its levels (-1 and 1) in different flasks (f1-f16).
Extraction and LC-DAD analysis
The mycelium extraction was done by harvesting fungal mycelia by filtration on paper filter no. 11 and extracting it with 50 mL of ethanol. The aqueous phase was extracted in triplicate with 50 mL of ethyl acetate. The extracts were dried under reduced pressure, re-dissolved in 3 mL of methanol and analyzed by reversed-phase LC-DAD.
The samples were analyzed by a gradient HPLC method for the detection of the naringin metabolites by injecting 10 μL of sample. The column used was a PHENOMENEX Luna Phenyl-hexyl 5 μ 250 × 4.6 mm. The mobile-phase composition consisted of water with 0.1% of formic acid (A) and methanol with 0.1% of formic acid (B) pumped at 1.0 mL.min-1. The mobile phase varied from 20% B in a linear gradient of 20 to 50% B over 10 minutes, followed by a linear gradient of 50% to 100% B in 20 minutes. After 10 minutes in 100% B, the gradient shifts to 20%B and stay in 20% B until next analysis. Naringin and its metabolites were detected at the wavelength of 289 nm and the data analysis was performed by CLASS-VP software.
LC-MS/MS analyses
HPLC-ESI/HRMS analyses were carried out on a LTQ-Orbitrap Thermo Fisher Scientific mass spectrometry system (Bremen, Germany) with the resolution set at 60K and operating in negative scan mode from 100-1500 Da. HPLC was fitted with a PHENOMENEX Luna Phenyl-Hexyl 5 μ 250 × 4.6 mm and samples were eluted using a linear gradient elution with acetonitrile and water from 20 to 100% with a 1.0 mL.min-1 flow.
Results and Discussion
Compounds identification
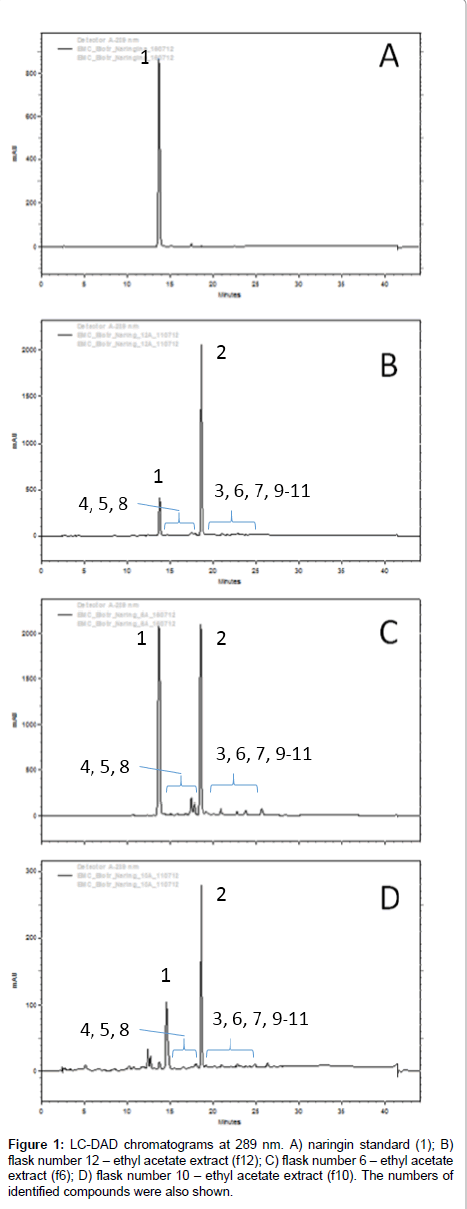
After extraction, the extracts were analyzed by LC-DAD and several chromatographic peaks were detected and exhibiting similar UV profile of naringin (1). The analyses and comparison with standard compounds indicated that part of 1 remained in all extracts and that the major biotransformation product was naringenin (2) by comparison with standards, while several candidate products were identified, as shown in Figure 1. HRMS analyses were performed with tandem mass spectrometry experiments in order to identify their chemical structures.
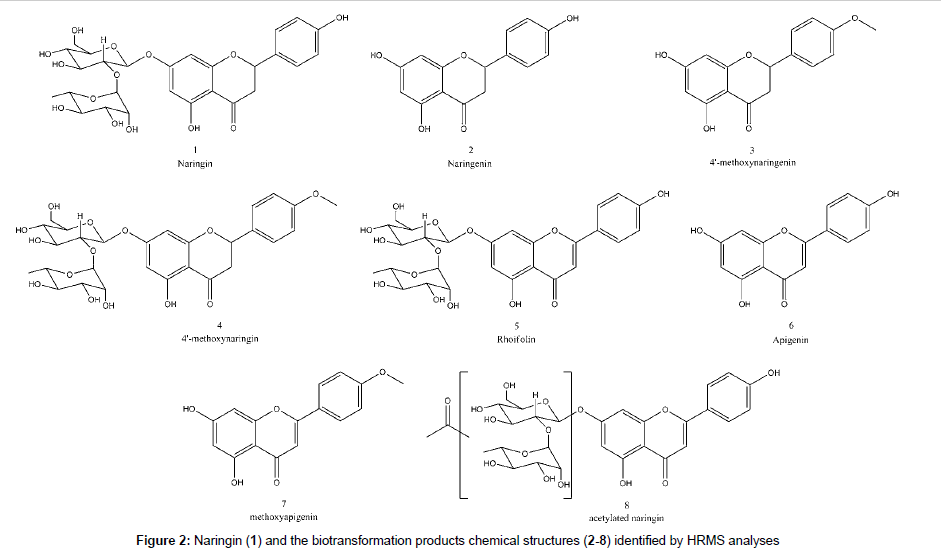
The peaks next to 1 and 2 showed similar UV pattern indicating them as possible biotransformation products. Their identification was achieved by HRMS analyses and comparison with literature information, according the discussions below. All identified compounds, which are shown in Figure 2, were not detected in control experiments, with fungus growing in the absence of substrate, and also in the medium with substrate but without fungus.
Naringin (1) fragmentation pattern is very characteristic due to C-ring retro Diels-Alder cleavage and sugar loss. The ethyl acetate extract from flask 6 of the designed experiment was utilized for the mass spectrometry analyses interpretation and identification of compounds; since it contain all biotransformation products detected at others extracts.
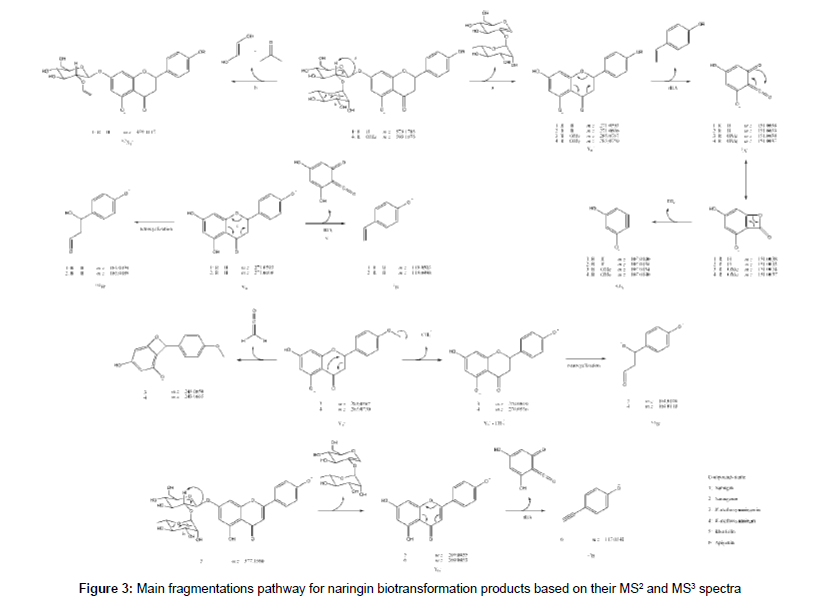
The HRMS ESI mass spectrum of 1 in negative mode shows a deprotonated molecule at m/z 579.1715 ([M-H]-) corresponding to molecular formula C27H31O14-. MS2 experiment led to m/z 271.0593 which corresponds to Y0- fragment, the same as deprotonated naringenin (2), according flavonoid fragmentation nomenclature states [21,22]. This was confirmed by MS3 experiments of m/z 271.0593, which led to m/z 151.0038 (C7H3O4-) and m/z 119.0498 (C8H7O-), 1,3A and 1,3B- fragments from C-ring retro Diels-Alder. It was also detected the ion at m/z 165.0194 (C8H5O4-) correspondent to a retrocyclization (0,4B- ion) and m/z 459.1117 (C19H23O13-) that is a 0,2X1 - fragment from sugar fragmentation. Table 2 summarizes the HRMS data for the detected compounds and the proposed fragmentation pathway for all the discussed structures is shown at Figure 3.
| Entry | Name | Deprotonated molecule | HRMS | MSn fragments | |
|---|---|---|---|---|---|
| Exp. | Δppm | ||||
| 1 | Naringin | C27H31O14- | 579.1715 | -1.1 | 459.1117 (C19H23O13-), 271.0593 (C15H11O5-), MS3: 165.0194 (C8H5O4-), 151.0038 (C7H3O4-),119.0503 (C8H7O-), 107.0140 (C6H3O2-) |
| 2 | Naringenin | C15H11O5- | 271.0606 | -1.9 | 165.0189 (C8H5O4-), 151.0033 (C7H3O4-), 119.0498 (C8H7O-), 107.0136 (C6H3O2-) |
| 3 | 4'-methoxynaringenin | C16H13O5- | 285.0767 | -0.7 | 270.0519 (C15H10O5-) / 243.0650 (C14H11O4-) / 164.0108 (C8H4O4-) / 151.0030 (C7H3O4-) / 107.0134 (C6H3O2-) |
| 4 | 4'-methoxynaringin | C28H33O14- | 593.1873 | -0.5 | 285.0750 (C16H13O5-), MS3: 270.0536 (C15H10O5-) / 243.0665 (C14H11O4-) / 164.0118 (C8H4O4-) / 151.0037 (C7H3O4-) / 107.0140 (C6H3O2-) |
| 5 | Rhoifolin | C27H29O14- | 577.1560 | -0.5 | 269.0435 (C15H9O5-) |
| 6 | Apigenin | C15H9O5- | 269.0453 | -0.7 | 117.0341 (C8H5O-) |
| 7 | Methoxyapigenin | C16H11O5- | 283.0606 | -2.1 | HRMS fragments not detected |
| 8 | Acetylated naringin | C29H33O15- | 621.1818 | 0.2 | 579.1689 (C27H31O14-) / 561.1585 (C27H29O13-) / 501.1225 (C25H25O11-) / 271.0596 (C15H11O5-) |
Table 2: HRMS Full Scan and MSn data for naringin (1) and biotransformation products (2-8)
The spectrum of 2, previously identified with comparison to standard as naringenin in LC-DAD, exhibited a deprotonated molecule at m/z 271.0606 correspondent to C15H11O5-. The MS2 spectrum produced ions at m/z 165.0189 (C8H5O4-), m/z 151.0033 (C7H3O4-), m/z 119.0498 (C8H7O-) and m/z 107.0136 (C6H3O2-) corresponding to 0,4B-, 1,3A-, 1,3B- and 0,4A-, respectively.
The mass spectrum of 3 showed a deprotonated molecule at m/z 285.0767 (C16H13O5-), an addition of 14 Da to 2, indicating a methylation. The fragment at m/z 270.0519 (C15H10O5-) mean a loss of methyl radical, confirming the methylation and fragments at m/z 243.0650 (C14H11O4-) indicate ketene loss (42 Da) from deprotonated molecule, previously reported for naringin fragmentation [23]. Ions with m/z 151.0030 and 107.0134 corresponding to 1,3A- and 0,4A- part of molecule confirms that the methyl moiety is located at 4´ position at B ring, since O-methoxylated fragment produced is neutral. There is one fragment at m/z 164.0108 (C8H4O4-) corresponding to ketene loss radical ion 0,4B-.. Thus, 3 is identified as 4’-methoxynaringenin.
The mass spectrum of 4 indicates a 14 Da addition, methylation, to naringin, since deprotonated ion at m/z 593.1873 correspond to C28H33O14. This modification was confirmed to be in position 4´ because the MS2 showed a fragment at m/z 285.0750 similar to 3 and MS3 of this ion showed the same fragmentation pattern, confirming 4 as 4’-methoxynaringin.
A loss of 2 Da in comparison to 1 was detected in the full scan spectrum of 5, indicating a loss of two hydrogens. The m/z 577.1560 was correlated to the molecular formula C27H29O14- and the fragmentation produced the m/z 269.0435 ion (C15H9O5-) which corresponds to a dehydrogenation at C ring in 2. In that way, 5 could be identified as rhoifolin.
The spectrum for the next compound (6) shows a deprotonated molecule at m/z 269.0453 (C15H9O5-), which suggest to be apigenin, the rhoifolin aglycone. The MS2 confirmed the proposal, since it produced the 1,3B- fragment at 117.0341 (C8H5O-). The addition of a methyl (14 Da) to 6 gave the 7 mass spectrum with a deprotonated molecule at m/z 283.0606 (C16H11O5-), which corresponds to methoxyapigenin, but no fragment was detected in MS2 experiments. Low resolution mass spectrometry collision-induced dissociation experiments evidences methyl position in the molecule at A ring (at position 4 or 6) as m/z 119 (1,3B-) and m/z 163 (1,3A-) ions were observed.
One acylated biotransformation product of 1 was detected (8) with m/z 621.1818 (C29H33O15-), corresponding to an addition of 42 Da. The MS2 experiments exhibit the ions at m/z 579.1689 (C27H31O14-), m/z 561.1585 (C27H29O13-), m/z 501.1225 (C25H25O11-) and m/z 271.0596 (C15H11O5-), corresponding to loss of ketene, water from deprotonated naringin, acetic acid and sugar, respectively. Due to 42 and 60 Da loss from parent ion, the acylation is located at saccharide moiety, but the exact position of acetylation in the saccharide could not be determined using only mass spectrometry.
Some unexpected deprotonated molecules with m/z 553.1143 (C31H21O10-), m/z 555.1291 (C31H23O10-), m/z 839.1973 (C47H35O15-) and m/z 863.2397 (C43H43O19-) were detected (9, 10, 11 and 12, respectively). They exhibited a similar fragmentation pattern, suggesting, for example, 9 and 10 to be a pair of biflavonoid composed of 2 and 7 due to MS2 ions at m/z 271.0595 (C15H11O5-) and m/z 283.0595 (C16H11O5-). These fragments were confirmed by MS3 data at m/z 151.0040 (C7H3O4-) and m/z 163.0040 (C8H3O4-), respectively. The mass spectrum of 11 present m/z 555.1256 (C31H23O10-) and m/z 283.0589 (C16H11O5-) as MS2 fragments, indicating a coupling of 7 and 9 molecules, as MS3 experiment for m/z 555.1256 (C31H23O10-) confirmed this as the same ion as 9 due to m/z 283.0616 (C16H11O5-) and m/z 271.0616 (C15H11O5-). Therefore this compound was suggested to be a triflavonoid. Accordingly, 12 was suggested to be another triflavonoid as result of coupling 7 to 1, due its MS2 spectrum exhibits an ion at m/z 579.1675 (C27H31O14-). This is a very interesting result, as biflavonoids and triflavonoids were not reported as biotransformation products in the literature, being the first report of this event.
Fractional Factorial design
It is possible to verify that different parameters for culture conditions affect directly biotransformation of naringin by M. inaequisporus, similar as proposed by the OSMAC methodology for secondary metabolism [24]. During the experimental part 11 compounds were observed in the total ion chromatogram of extracts. The relative area of each compound was recorded (as symbols “-” for undetected, “+”, “++” and “+++” for relative areas of increasing relative concentration) and our intention was to identify a condition that is able to produce more compounds as shown in Table 3. In addition, our main interest is also to identify an experiment that produces more rhoifolin, apigenin and methoxyapigenin. The biflavonoids and triflavonoids were not accounted for this calculation.
In this case, weights were attributed for each response: naringin, for example, received a weight 1, naringenin was 2, 4-methoxynaringenin and 4- methoxynaringin were 3 and the three most important compounds mentioned before received a weight 4. After that, all 8 responses for each experiment were combined using geometric mean. This response was used to calculate the effects and to identify the most important variables. The fractional factorial design permits to identify the factors that enhance the biotransformation capability of the microorganism. The most influent factors are in this order, luminosity (variable 6), carbon source (variable 1) and shaking (variable 5) as seen for flasks 6 and 12. The best responses were obtained when the flasks were maintained in the dark (level -1 for variable 6), dextrose was used (level +1 for variable 1) and static (level -1 for variable 5). The other variables (2, 3, 4, 7 and 8) did not present remarkable effects in the studied levels and can be fixed in the most suitable condition.
Mucor inaequisporus biotransformation capability
The Mucor is the biggest genus from Mucorales order and comprises in the major part of saprophytic microorganisms that causes food ripening and grain contamination. Some few species are mammalian and plant pathogens [25]. One of the main biotechnology applications for Mucor genera is to biotransform natural products, especially steroidal like compounds and terpenoids [26,27]. On the other hand, there are few reports about biotransformation of flavonoids by Mucor genera. Mucor ramannianus can perform A- and B-ring hydroxylation, demethylation, sulfation and glycosylation in hesperetin [28], hydroxylation, sulfation and glycosylation in cannflavin A and B [29], glycosylation in kaempferol [30], B-ring hydroxylation, glycosylation, and carbonyl reduction in 4’-hydroxyflavanone [31]. Furthermore, Mucor hiemalis glycosylated 8-prenylnaringenin [32], Mucor spinosus performed the same reaction in cardamonin [33] and a Mucor miehei lipase was used to prepare 3-O-acylcatechins [34], demonstrating the enzymatic potential for flavonoid modification of this genera.
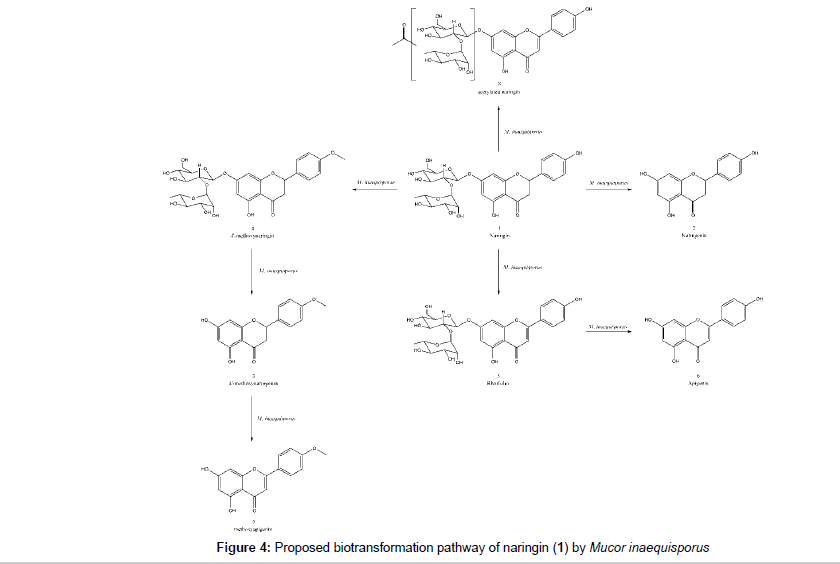
The LC-MS/MS analyses at high resolution allowed the identification of several microbial products as naringenin (2), 4’-methoxynaringenin (3), 4’-methoxynaringin (4), rhoifolin (5), apigenin (6), 4- or 6-methoxyapigenin (7) and a sugar acetylated naringin (8). Biflavonoids and triflavonoids were also detected in some extracts. There are no literature report for biflavonoids (9 and 10) and triflavonoids (11 and 12) as metabolites obtained from microbial transformation of flavonoids. The microorganism was able to perform deglycosylation, dehydrogenation and O-methylation whereas the common hydroxylation [28,29,31] and glycosylation [28-33] reaction for Mucor species was not observed. The proposed biotransformation pathway is represented at Figure 4. Naringin, the identified naringin metabolites and its qualitatives correlation in each flask are summarized in Table 3.
| Compound | f1 | f2 | f3 | f4 | f5 | f6 | f7 | f8 | f9 | f10 | f11 | f12 | f13 | f14 | f15 | f16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Naringin (substrate) | +++ | +++ | +++ | ++ | +++ | ++ | +++ | +++ | +++ | + | +++ | ++ | +++ | +++ | +++ | +++ |
| Naringenin | + | + | + | + | + | +++ | + | ++ | + | +++ | + | +++ | + | + | ++ | + |
| 4'-methoxynaringenin | - | - | + | - | + | ++ | - | + | - | + | - | ++ | - | + | + | - |
| 4'-methoxynaringin | ++ | ++ | ++ | ++ | ++ | + | ++ | ++ | ++ | + | ++ | ++ | ++ | ++ | ++ | ++ |
| Rhoifolin | ++ | ++ | ++ | + | ++ | ++ | ++ | ++ | ++ | - | ++ | + | ++ | ++ | ++ | ++ |
| Apigenin | + | - | + | - | - | ++ | + | + | - | + | - | ++ | - | + | ++ | - |
| Methoxyapigenin | - | - | - | - | - | ++ | - | + | - | + | - | ++ | - | - | + | + |
| Acetylated naringin | ++ | ++ | ++ | ++ | ++ | + | ++ | ++ | ++ | + | ++ | ++ | ++ | ++ | ++ | ++ |
Table 3: Qualitative correlation between naringin and identified biotransformation products in all experiment flasks where “+” means detected by MS, “++” means formation of a chromatographic band, “+++” means majority and “-” means undetected
Apigenin was once reported as biotransformation product from naringin [35] as well as acylated naringin, produced by Lipozyme catalysis from Thermomyces lanuginosus, so this is the first time a whole cell microorganism perform these reactions in naringin [36]. In contrast, 4’-methoxynaringin, 4’-methoxynaringenin, methoxyapigenin and rhoifolin were not previously reported as naringin biotransformation product in literature, so these novel results are very interesting for assessing the Mucor inaequisporus enzymatic potential.
Conclusion
The fungus Mucor inaequisporus was capable to perform the biotransformation of naringin to eleven different products which were identified through HRMSn analysis and extensive fragmentation studies. The great number and great structural diversity created by the fungus indicates the vast enzymatic machinery and therefore it inspires the use of this microorganism as a living catalyst that could be used as source for important structural modifications in other substrates and source of valuable enzymes. The mass spectrometry has showed to be a powerful technique for elucidation of flavonoid biotransformation products.
Acknowledgements
The authors are grateful to FAPESP for granting fellowship for Ph.D. study (2011/09580-4 for Enzo Monte Canedo and 2014/03510-2 for Taicia Pacheco Fill), CNPQ (Sisbiota-Brasil), and CAPES for research financial support.
References
- Yao HL, Jiang YM, SHI J, Tomás-Barberán FA, Datta N, et al. (2004) Flavonoids in food and their health benefits. Plant Food Hum Nutr 59: 113-122.
- Das S, Rosazza JPN (2006) Microbial and enzymatic transformations of flavonoids. J Nat Prod 69: 499-508.
- Sanchez-Gonzalez M, Rosazza JPN (2004) Microbial transformations of chalcones: hydroxylation, O-demethylation, and cyclization to flavanones. J Nat Prod 67: 553-558.
- Tsen HY, Yu GK (1991) Limonin and naringin removal from grapefruit juice with naringinase entrapped in cellulose triacetate fibers. J Food Sci 56: 31-34.
- Lee SJ, Kim JC, Kim MJ, Kitaoka M, Park CS, et al. (1999) Transglycosylation of naringin by Bacillus stearothermophilus maltogenic amylase to give glycosylated naringin. J Agric Food Chem 47: 3669-3674.
- Kontogianni A, Skouridou V, Sereti V, Stamatis H, Kolisis FN (2001) Regioselective acylation of flavonoids catalyzed by lipase in low toxicity media. Eur J Lipid Sci Tech 103: 655-660.
- Pedro HAL, Alfaia AJ, Marques J, Vila-Real HJ, Calado A, et al. (2007) Design of an immobilized enzyme system for naringin hydrolysis at high-pressure. Enzyme Microb Tech 40: 442-446.
- Ribeiro IAC, Ribeiro MHL (2008) Kinect modelling of naringin hydrosylis using a bitter sweet alfa-rhamnopyranosidase immobilized in k-carrageenan. J Mol Catal B Enzym 51: 10-18.
- Marques J, Vila-Real HJ, Alfaia AJ, Ribeiro MHL (2007) Modelling of the high pressure-temperature effects on naringin hydrolysis based on response surface methodology. Food Chem 105: 504-510.
- Chang HY, Lee YB, Bae HA, Huh JY, Nam SH, et al. (2011) Purification and characterisation of Aspergillus sojae naringinase: The production of prunin exhibiting markedly enhanced solubility with in vitro inhibition of HMG-CoA reductase. Food Chem 124: 234-241.
- Ni H, Xiao AF, Cai HN, Chen F, You Q, et al. (2012) Purification and characterization of Aspergillus niger a-L-rhamnosidase for the biotransformation of naringin to prunin.Afr J Microbiol Res 6: 5276-5284.
- Ciegler A, Lindenfelser LA, Nelson GEN (1971) Microbial transformation of flavonoids. Appl Microbiol 22: 974-979.
- Ye H, Xu H, Yu C, Dai Y, Liu G, et al. (2009) Hydroxylation of naringin by Trichoderma harzianumto dramatically improve its antioxidative activity. Enzyme Microb Tech 45: 282-287.
- Miyake Y, Minato K, Fukumoto S, Yamamoto K, Oya-Ito T, et al. (2003) New potent antioxidative hydroxyflavanones produced with Aspergillus saitoi from flavanone glycoside in citrus fruit. Biosci Biotechnol Biochem 67: 1443-1450.
- Cheng KJ, Krishnamurty HG, Jones GA, Simpson FJ (1971) Identification of products produced by the anaerobic degradation of naringin by Butyrivibrio sp. C3. Can J Microbiol 17: 129-131.
- Dumenil G, Cremieux A, Phan-Tan-Luu R, Combet M (1975) Bioconversion from DL-Homoserine to L-Threonine. Eur J Appl Microbiol Biotechnol 1: 213-220.
- Milanova R, Stoynov N, Moore M (1996) The optimization of triptoquinone production by Cunninghamella elegansusing factorial design. Enzyme Microb Tech 19: 86-93.
- Waché Y, Aguedo M, LeDall MT, Nicaud JM, Belin JM (2002) Optimization of Yarrowia lipolytica’s β-oxidation pathway for γ-decalactone production. J Mol Catal B Enzym 19-20: 347-351.
- Zhang XY, Peng Y, Su ZR, Chen QH, Ruan H, et al. (2013) Optimization of biotransformation from phytosterol to androstenedione by a mutant Mycobacterium neoaurum ZJUVN-08. J Zheijang Univ Sci B 14: 132-143.
- Santiago ALCMA, Rodrigues A, Canedo EM, Filho ER (2013) Taxonomic studies on Mucor inaequisporus, isolated for the first time in South America. Mycotaxon 124: 219-229.
- Ma YL, Li QM, Heuvel HV, Claeys M (1997) Characterization of flavone and flavonol aglycones by collision-induced dissociation tandem mass spectrometry. Rapid Commun Mass Sp 11: 1357-1364.
- Domon B, Costello CE (1988) A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate J 5: 397-409.
- Xu F, Liu Y, Zhang Z, Yang C, Tian Y (2009) Quasi-MSn identification of flavanone 7-glycoside isomers in Da Chengqi Tangby high performance liquid chromatography-tandem mass spectrometry. Chin Med 4: 15.
- Bode HB, Bethe B, Hfs R, Zeeck A (2002) Big effects from small changes: possible ways to explore nature’s chemical diversity. Chembiochem 3: 619-627.
- Frank W, Roester U, Scholer HJ (1974) Sphaerule formation by a Mucor species in the internal organs of Amphibia. Preliminary report. Zentralbl Bakteriol Orig A 226: 405-417.
- Silva EO, Furtado NAJC, Aleu J, Collado IG (2014) Non-terpenoid biotransformations by Mucor species. Phytochem Rev: Ahead of print.
- Silva EO, Furtado NAJC, Aleu J, Collado IG (2013) Terpenoid biotransformations by Mucor species. Phytochem Rev 12: 857-876.
- Herath W, Khan IA (2011) Microbial metabolism. Part 13. Metabolites of hesperetin. Bioorg Med Chem Lett 21: 5784-5786.
- Ibrahim AK, Radwan MM, Ahmed SA, Slade D, Ross SA, et al. (2010) Microbial metabolism of cannflavin A and B isolated from Cannabis sativa. Phytochemistry 71: 1014-1019.
- Ibrahim A, Khalifa SI, Khafagi I, Youssef DT, Khan S, et al. (2008) Microbial metabolism of biologically active secondary metabolites from Nerium Oleander L. Chem Pharm Bull (Tokyo) 56: 1253-1258.
- Mikell JR, Herath W, Khan IA (2011) Microbial metabolism. Part 12. Isolation, characterization and bioactivity evaluation of eighteen microbial metabolites of 4'-hydroxyflavanone. Chem Pharm Bull (Tokyo) 59: 692-697.
- Kim HJ, Kim SH, Kang BY, Lee IS (2008) Microbial metabolites of 8-prenylnaringenin, an estrogenic prenylflavanone. Arch Pharm Res 31: 1241-1246.
- Xu J, Yang L, Zhao SJ, Chou GX, Wang ZT (2011) Microbial glycosylation of cardamonin by Mucor spinosus. Yao Xue Xue Bao 46: 733-737.
- Patti A, Piatelli M, Nicolosi G (2000) Use of Mucor miehei lipase in the preparation of long chain 3-O-acylcatechins. J Mol Catal B Enzym 10: 577-582.
- Kostrezewa-Suslow E, Dmochowska-Gladysz J, Janeczko T (2010) Microbial transformation of selected flavanones as a method of increasing the antioxidante properties. Z Naturforsch C 65: 55-60.
- Luo XP, Du LH, He F, Zhou CH (2013) Controllable regioselective acylation of flavonoids catalyzed by lipase in microreactors. J Carbohydr Chem 32: 450-462.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 15199
- [From(publication date):
specialissue-2014 - Apr 03, 2025] - Breakdown by view type
- HTML page views : 10539
- PDF downloads : 4660