Enhancement of Drought Tolerance in Rice through Introgressiovn of Arabidopsis DREB1A through Transgenic Approach
Received: 04-Feb-2019 / Accepted Date: 28-Mar-2019 / Published Date: 08-Apr-2019
Abstract
With global temperatures on the raise, drought stress is turning out to be the major impediment to increase in rice production all over the world. In India, where 60% of the rice lands are rain-fed, rice production is severely hampered by drought as the crop depends heavily on monsoon rains. In this context, we attempted to incorporate tolerance against drought in Indica rice through introgression of AtDREB1A gene from Arabidopsis employing Agrobacterium mediated transgenic approach. Putative transgenics were generated and the presence of the incorporated gene was confirmed through Polymerase Chain Reaction (PCR) and Southern blotting assays. The inheritance pattern of the incorporated gene was Mendelian. The physiological studies conducted on eight transgenic lines revealed that introduced gene could confer significantly higher levels of tolerance to drought stress. This promising result suggests that transgenic approach can be a viable option for genetic enhancement of rice against abiotic stresses like drought.
Keywords: Transgenic; Indica rice; Pusa Basmati 1; Drought tolerance
Introduction
Global temperatures increased drastically in the last decade and continuous global warming prevailed across the globe. In India, highest temperatures were recorded according to the World Meteorological Organization report. Cereal production in Asian continent is severely affected by drought, predominantly during dry season. According to a recent report from Intergovernmental Panel on Climate Change, about one fifth area which corresponds to 23 million hectares of rice ecosystem is prone to drought. In eastern India, about 11% of the total agricultural GDP is affected by drought [1].
Indian agriculture is mainly monsoon based, with over 60% of agricultural land being rain-fed, a failed monsoon causes a crippling impact on food production and farmers often encounter a complete loss of crops thus threatening their livelihoods and food security. As there is no control over the monsoon, it is essential to develop rice varieties that can grow in adverse situations like drought. Rice is mostly grown in hydrological conditions and limited success for drought tolerance was achieved through conventional approaches due to the lack of available donors in the rice gene pool to improve drought tolerance, which is a complex polygenic trait. Drought is fast becoming the most serious abiotic stress due to increase in temperatures and decrease in ground water levels.
Recent progress on targeting abiotic stress tolerance focused mainly on structural and regulatory genes through genetic engineering approach. This study focused on the applications of regulatory genes including: i) Transcription factor genes and ii) Dehydration Responsive Element Binding Factors (DREBs). It was reported that there was an increase in level of transcription of several downstream target genes at a single stretch by over expressing certain transcription factors [2]. Yamaguchi and Shinozaki [3] first reported the presence of transcription factor termed as Dehydration-Responsive Element (DRE), in the promoter region of RD29A gene in Arabidopsis. DRE binds to special signature sequence and targets the expressions of several downstream genes. Specific signature sequence of C-repeats in DRE transcription factor, are termed as CRT elements or CRT-binding factors. Moreover, these Dehydration Responsive Element-Binding (DREB) factors were found to be targeting several genes involved in drought, and cold tolerance and they function independent of leaf senescence hormone abscisic acid.
Experiments carried out in the model plant Arabidopsis have shown that the constitutive over expression of either DREB1A from rice (OsDREB1A) or DREB1A from maize (ZmDREB1A) have resulted in the regulation of downstream targeted genes imparting dehydration tolerance [4], cold and salinity tolerance [5]. Successful results of desiccation tolerance in Arabidopsis had encouraged various research groups to target AtDREB1A gene into various crops but the results varied under the influence of constitutive promoters (e.g. 35S, ZmUbi, OsActin1) and stress inducible promoters (e.g. RD29A, OsHVA22p). Desiccation tolerance was more profound in the case of stress inducible promoters [6] than the constitutive promoters [7].
Considering the success with DREB genes, the present work, aimed to generate transgenic rice through incorporation of AtDREB1A transcription factor under the control of RD29A promoter into Pusa Basmati 1 (PB1), an elite Basmati rice variety, which is extensively grown in the north eastern region of India with an yield potential of 4.5 t/ha and is amenable to tissue culture [8].
Materials and Methods
Generation of transgenic plants
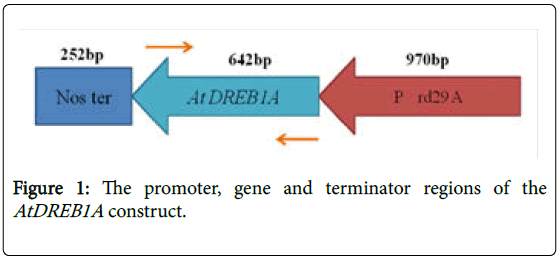
Mature kernels of elite Indica rice genotype Pusa Basmati 1 were surface sterilized [9] and inoculated on MS media [10] supplemented with 2,4-Dichlorophenoxy acetic acid (2,4-D @2.0 mg L-1) and incubated at 26 ± 1°C in dark for three weeks. Scutellum derived embryogenic callus was used as the target tissue for Agrobacterium transformation using a super virulent Agrobacterium tumefaciens strain LBA4404 (pSB1) carrying the binary vector pC1200-RD29AAtDREB1A with a hygromycin-resistant gene (HPT) as plant selection marker and chloramphenicol as the bacterial selection marker. The target gene was driven by the stress-inducible RD29A promoter and a nos terminator derived from Arabidopsis thaliana (Figure 1).
Agrobacterium mediated transformation, co-cultivation, washing, selection (four cycles of 15 days each) were carried out as per Saikrishna et al. [11] and the resistant calli were transferred onto regeneration media as per Chaitanya et al. [8]. Shoots were transferred into rooting media (MS media supplemented with maltose (50 g l-1), naphthalene acetic-acid (1.0 mg L-1), and kinetin (0.1 mg L-1)). Putative transgenic plants with well-established roots were transferred into plastic pots having sterile soil in the institutional biosafety glass house.
Molecular analyses (polymerase chain reaction (PCR) and Southern blot analysis)
DNA was extracted from the fresh leaves employing CTAB method as proposed by Murray and Thompson [12]. Isolated DNA was examined for the presence of transgene (DREB1A) and antibiotic marker gene (HPT) with the help of specific primers through Polymerase Chain Reaction (PCR) assays in a total reaction volume of 10 μL per reaction in a programmable thermal cycler (Eppendorf Vapo protect). The constituents of the PCR reaction include: 1 μL of plant DNA (20 ng), 0.8 μL of 2.5 mM dNTPs (Fermentas), 1.0 μL of 10X PCR buffer (Sigma), 0.2 μL of Taq DNA Polymerase (5 U/μL Sigma), 1 μL each of both forward & reverse primers (5 pico moles/μL Sigma) and 5 μL of autoclaved sterile distilled water. The PCR reaction conditions followed for both the genes include; an initial denaturation of template DNA at 94°C for 5 min followed by 40 repeated cycles of denaturation at 94°C for 1 min, primer annealing at 58°C for 1 min, and primer extension at 72°C for 1 min followed by a final extension for 5 min. PCR amplicons were separated through electrophoresis on 1.2% agarose gel containing 0.5 μg/mL ethidium bromide (in 1X TBE electrophoresis buffer). Separated PCR products were photographed in a gel documentation system to examine the size of the product compared with one-kb molecular weight marker.
For Southern analysis, DNA isolated from all the test samples and control were subjected to overnight restriction digestion at 37°C with Hind III (Promega) restriction enzyme for gene of interest DREB1A in a total reaction volume of 40 μL. Restriction digestion was checked by loading one microliter each of digested sample on to 0.8% (w/v) agarose gel, pre-stained with ethidium bromide (0.5 μg/mL) and observed under UV. Rest of the digested samples were fractionated on 0.8% (w/v) agarose gel. The gel was depurinated and equilibrated for 30 min each subsequently with respective buffers and the DNA fragments were transferred from the gel to Hybond-N+ nylon membrane under alkaline denaturation conditions according to manufacturer’s (Amersham, Arlington Heights, IL) instructions. The membrane was pre-hybridized with 50 mL of hybridization buffer for at least 1 h according to Zoller and Smith [13] in to which sheared single stranded salmon sperm DNA (10 μg/mL) was added for blocking. DREB1A fragment (0.56 kb) to be used as hybridization probe was radio labelled with (α-P32) dCTP using the Rediprime Labeling Kit (Promega). After the overnight hybridization, the membranes were washed thrice for 30 min each subsequently with wash buffers 1,2,3 as proposed by Sambrook and Russell [14]. Excess wash solution was removed and the membrane was wrapped with saran-wrap and auto-radiographed overnight depending on the radioactive count by a GM counter and visualized under a phosphorimager (Typoon-5000 GE-Health care).
RT-PCR
Total RNA was extracted from the leaves of both stress imposed Southern positive transgenic lines as well as non- transformed control plants and RT-PCR was performed as per the standard protocol followed in our lab according to Rao et al. [15]. Primers used for DREB1A gene amplification were same as mentioned for PCR analysis.
Physiological Studies
To study the physiological parameters, transgenic rice lines of T3 generation were grown in plastic pots in triplicates, containing 5 kg of soil each. To maintain uniformity in the soil texture, soil was dried, finely ground and mixed thoroughly. Four to five seeds were germinated in each pot and after a week, a single healthy seedling was maintained per each pot. Drought stress experiments were conducted as per Liu et al. [16] and a ten day stress was imposed on the twentyone day old seedlings by withdrawing irrigation and observations on different physiological and biochemical parameters were taken at two days interval from the day of imposition of stress.
Relative water content
Relative Water Content (RWC) was calculated as per formula given by Bonnet et al. [17]. Three kinds of weight viz., Fresh Weight (FW), Turgid Weight (TW) and Dry Weight (DW) were measured subjecting the leaves to three different conditions. Fresh weight was determined from the central portions of the fully expanded leaf samples collected during midday immediately after collection. These leaves were maintained in water for turgidity in closed petri-dish for 8 h under normal room temperature, blotted to remove excess surface moisture and weighed to determine turgid weight. Later, the samples were oven dried for 24 h at 80°C and weighed to determine dry weight.
Relative chlorophyll content
Chlorophyll concentration in rice leaves was determined using the chlorophyll meter as described by Markwell et al. [18] and such values have been shown to be positively correlated (r2=0.93) with destructive chlorophyll measurements in rice [19].
Soil moisture content (SMC)
At the end of the stress period, soil moisture content was measured from a depth of 15 cm to 30 cm soil according to Brady [20].
Proline content
Proline content in the test sample from the standard curve was calculated using the following formula given by Bates et al. [21].
Gas exchange parameters
Gas exchange parameters were measured from the top second leaf in comparison to the standard parameters using a portable infrared gas analyzer according to Nataraja and Jacob [22]. Instantaneous Water Use Efficiency (WUE) is calculated as the ratio between net photosynthetic rate (A) and transpiration rate (T).
Chlorophyll fluorescence
Chlorophyll fluorescence was measured as given by Panda and Sarkar [23].
Results
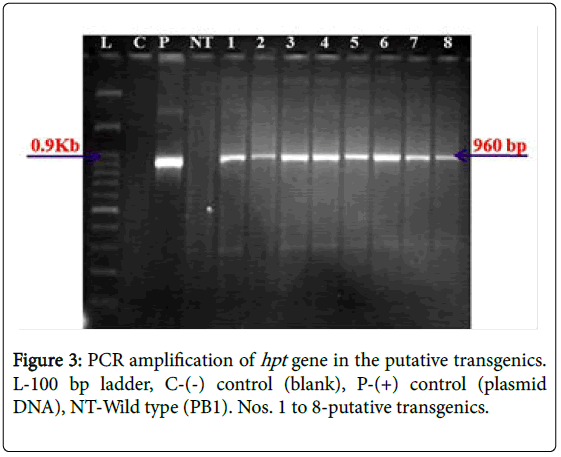
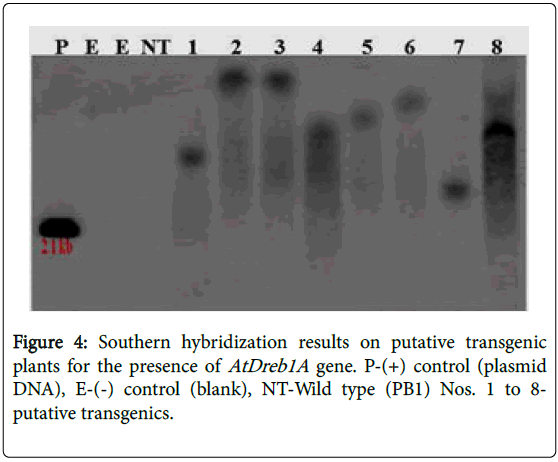
A total of 72 putative transgenic plants were obtained and grown in the transgenic greenhouse till maturity. PCR assays, conducted could amplify the 560 bp fragment precise to the introduced target gene (AtDREB1A ) and a 960 bp fragment specific to the selectable marker ( gene) in all the plants (Figures 2 and 3). Further studies were conducted with eight plants that were generated from different calli selected during the selection cycles. Southern hybridization studies, conducted using a 560 bp radioactive gene sequence as the probe, established the incorporation of the AtDREB1A gene in the rice genome. The presence of a strong signal in all the plants (D1-8) confirms the clear integration of the target gene (AtDREB1A gene) into the rice genome while hybridization signal was not detected in the non-transformed (wild type) plants (Figure 4). The signal pattern in the Southern blots was different in all the test plants, except for the plants 2 & 3 which appeared to be similar. The size of the hybridized fragment varied from plant to plant while the expected size of the fragment in the control (plasmid DNA) is 2.1 kb, suggesting the presence of at least seven independent events.
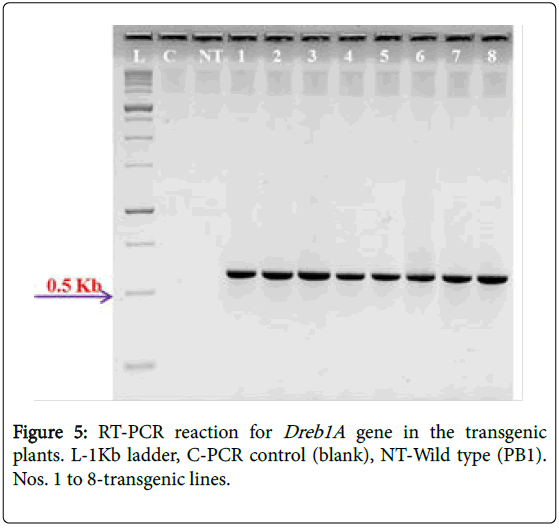
Expression of the introduced gene in the transformed plants was checked by reverse transcription polymerase chain reaction. The results suggest that in all the eight putative transgenics, the expression of introduced gene was observed (Figure 5). All these molecular analyses were performed on plants of T0 generation (putative transgenics) and studies continued to estimate the transmission rates through PCR assays and it was observed to be in Mendelian proportion.
Physiological studies
Physiological studies were conducted on T4 generation lines and homozygous lines of each transgenic were grown for physiological studies to assess the lines for their level of drought tolerance with nontransformed PB1 plants as controls (Figure 6).
Relative water content (RWC)
During unstressed treatment there was not any practical difference in the leaf RWC between control and transgenic lines and was in the range of 84% to 93% while during stress (10 d), the RWC of both transformed and non-transformed leaves dropped quickly from 90.5% to 53.2%, in control, whereas the proportion of RWC in most of the transgenic plants were higher than in wild type which was of the following order: D-1(68.8%), D-2(62.1%), D-3(72.6%), D-4 (74.3%), D-5 (64.3%), D-6 (61.7%), D-7(65.2%) and D-8 (72.4%). After 10 days of dehydration stress, the RWC of the transgenic lines had just decreased by 11.6% to 34.7% in comparison to 37.3% as in case of control plants. However, the rate of decrease of RWC was less in lines such as D-6, D-2, D-5, D-7 and D-1 in comparison to lines, D-8, D-3 and D-4 (Table 1).
| Percentage of Relative Water Content | Relative chlorophyll content | Proline content (μ mole/gm fresh wt.) | SMC | Drought score | Recovery score | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6-DAS | 10-DAS | 6-DAS | 10-DAS | 6-DAS | 10-DAS | 10-DAS | |||||||||
| WW | WS | WW | WS | WW | WS | WW | WS | WW | WS | WW | WS | ||||
| PB1 | 92 | 59.8 | 90.5 | 53.2 | 32.3 | 28.2 | 32.2 | 27.1 | 17.7 | 46.5 | 17.2 | 140.6 | 11.36 | 9 | 9 |
| D1 | 86.7 | 72.7 | 86.7 | 68.8 | 31.3 | 25.2 | 31.6 | 28.4 | 16.5 | 167.5 | 19.4 | 368.9 | 10.76 | 3 | 1 |
| D2 | 91.7 | 75.9 | 91.4 | 62.1 | 30.6 | 29.4 | 30.4 | 29.8 | 19.3 | 119.8 | 19.2 | 277.1 | 9.12 | 1 | 1 |
| D3 | 93 | 78.7 | 84.2 | 72.6 | 31.5 | 30.7 | 31.7 | 30.4 | 15.2 | 106.8 | 18.7 | 251.4 | 10.23 | 3 | 1 |
| D4 | 91.3 | 67.2 | 91.2 | 74.3 | 34.8 | 27.6 | 33.2 | 30.5 | 15.8 | 216.2 | 21.1 | 407.2 | 11.05 | 3 | 1 |
| D5 | 85.7 | 63.1 | 86.4 | 64.3 | 33.3 | 30.9 | 32 | 30.6 | 17.4 | 211.8 | 20.5 | 405.9 | 10.2 | 1 | 1 |
| D6 | 90.5 | 68.3 | 96.4 | 61.7 | 34.2 | 31.5 | 33 | 30.8 | 22.4 | 172.5 | 18.4 | 391.9 | 10.05 | 3 | 1 |
| D7 | 84 | 67.4 | 83.3 | 65.2 | 42.4 | 40.7 | 41.2 | 39.5 | 35.1 | 186.1 | 17.8 | 396.2 | 10.29 | 3 | 1 |
| D8 | 86.7 | 78.1 | 84 | 72.4 | 41.4 | 39.7 | 40.7 | 37.7 | 41.5 | 145.3 | 21.8 | 347.8 | 10.92 | 1 | 1 |
Table 1: Mean data table of physiological and biochemical characterization. DAS-days after stress
Chlorophyll content
SPAD values of the transgenic and non-transgenic control plants were almost similar, and ranged from 32.3 to 42.4 before stress but after initiation of stress, the chlorophyll values of the stressed plants decreased ranging from 25.2 to 40.7 with most of the transgenic plants retaining 28.4% to 39.5% of the chlorophyll content on the 10th day of stress. However, after re-irrigation for 2 days, i.e., 12th day, increased levels of chlorophyll were observed in transgenic plants while such increase was not observed in control.
Proline content
During stress treatment, the levels of proline increased both in transgenic and control plants and it was found to be higher after 6 days in transgenic lines as follows: D-4 (216.2 μM/g fr wt.), D-5 (211.8 μM/g fr wt.), D-7 (186.1 μM/g fr wt.), D-6 (172.5μM/g fr wt.) and D-8 (145.3 μM/g fr wt.), but after 10 days of stress, proline content was highest as observed in D-4 (407.2 μM/g fr wt) followed by D-5 (405.9 μM/g fr wt), D-7 (396.2 μM/g fr wt), D-6 (391.9 μM/g fr wt) and D-1 (368.9 μM/g fr wt) whereas the unstressed plants showed the lower levels of proline ranging from 15.2 μM/g fr wt to 41.5 μM/g fr wt (Table 1), while there was not any difference in the proline levels of unstressed transgenic and control plants.
Photosynthetic gas exchange parameters
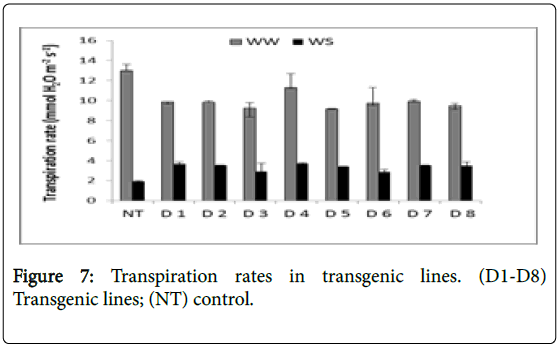
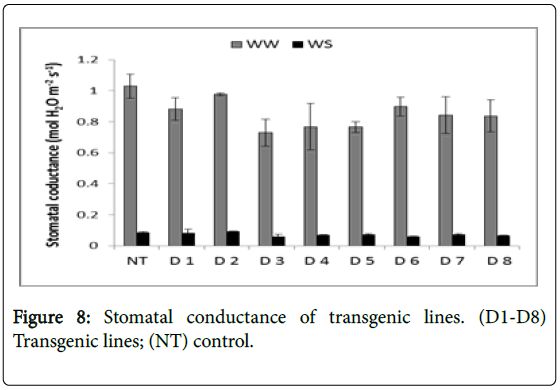
Rate of transpiration, stomatal conductance, and instantaneous water use efficiency of the transgenic lines (D1-D8) were measured in comparison to Non-Transgenic line (NT) under normal Well Water (WW) and Water Stress (WS) conditions (Table 2). Transpiration rates were in the range of 9 to 13 (mM H2O m-2s-1) under Well Watered (WW) condition but decreased to 1.9 to 3.6 (mM H2O m-2s-1) under Water Stress (WS) conditions. Transpiration rates in transgenic lines were more compared to control with D4 showing highest transpiration rate (3.6) followed by D1 (3.64), D2 (3.52), D7 (3.5), D8 (3.49), D3 (2.88) and D6 (2.87) in the decreasing order (Figure 7). Stomatal conductance ranged from 0.72 to 1.03 (M H2O m-2s-1) under well watered condition but decreased within the range from 0.06 to 0.09 (M H2O m-2s-1) under water stress conditions (Figure 8).
| S.N. | Transpiration rate | Stomatal Conductivity | Water Use Efficiency | Fv/Fm | Photosynthetic rate | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| WW | WS | WW | WS | WW | WS | WW | WS | WW | WS | |
| NT | 13.00 ± 0.59 | 1.93 ± 0.05 | 1.03 ± 0.07 | 0.08 ± 0.00 | 1.52 ± 0.02 | 1.74 ± 0.11 | 0.82 ± 0.05 | 0.70 ± 0.01 | 19.85 ± 0.71 | 03.37 ± 0.13 |
| D1 | 09.87 ± 0.00 | 3.64 ± 0.23 | 0.88 ± 0.07 | 0.08 ± 0.02 | 2.21 ± 0.00 | 2.85 ± 0.15 | 0.81 ± 0.02 | 0.75 ± 0.05 | 21.87 ± 0.00 | 10.39 ± 0.12 |
| D2 | 09.83 ± 0.11 | 3.52 ± 0.03 | 0.97 ± 0.00 | 0.09 ± 0.00 | 2.1 ± 0.06 | 2.91 ± 0.04 | 0.82 ± 0.03 | 0.75 ± 0.07 | 21.05 ± 0.86 | 10.27 ± 0.07 |
| D3 | 09.23 ± 0.57 | 2.88 ± 0.86 | 0.72 ± 0.08 | 0.06 ± 0.01 | 2.45 ± 0.04 | 3.11 ± 0.57 | 0.81 ± 0.03 | 0.74 ± 0.01 | 22.62 ± 0.98 | 08.47 ± 1.01 |
| D4 | 11.28 ± 1.40 | 3.68 ± 0.07 | 0.76 ± 0.15 | 0.06 ± 0.00 | 1.95 ± 0.35 | 2.62 ± 0.00 | 0.82 ± 0.06 | 0.74 ± 0.00 | 21.52 ± 1.27 | 09.67 ± 0.21 |
| D5 | 09.18 ± 0.02 | 3.38 ± 0.05 | 0.76 ± 0.03 | 0.07 ± 0.00 | 2.49 ± 0.00 | 3.22 ± 0.03 | 0.81 ± 0.05 | 0.75 ± 0.01 | 22.88 ± 0.10 | 10.94 ± 0.24 |
| D6 | 09.72 ± 1.56 | 2.87 ± 0.22 | 0.89 ± 0.06 | 0.06 ± 0.00 | 2.46 ± 0.54 | 3.90 ± 0.02 | 0.81 ± 0.02 | 0.76 ± 0.02 | 23.10 ± 1.36 | 11.21 ± 0.81 |
| D7 | 09.90 ± 0.22 | 3.50 ± 0.04 | 0.84 ± 0.11 | 0.07 ± 0.00 | 2.32 ± 0.08 | 3.55 ± 0.18 | 0.82 ± 0.01 | 0.76 ± 0.01 | 23.06 ± 1.31 | 12.45 ± 0.48 |
| D8 | 09.48 ± 0.21 | 3.49 ± 0.37 | 0.83 ± 0.10 | 0.06 ± 0.00 | 2.22 ± 0.02 | 3.39 ± 0.11 | 0.81 ± 0.05 | 0.74 ± 0.01 | 21.12 ± 0.63 | 11.83 ± 0.88 |
Table 2: Mean data table of photosynthetic studies in transgenic lines.
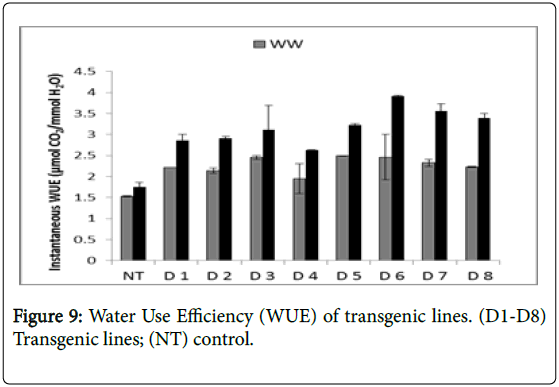
Water use efficiency ranged from 1.95 to 2.49 μM CO2/mM H2O under well watered condition but decreased within the range from 1.74 to 3.9 μM CO2/mM H2O under stress conditions. Water use efficiency rates in transgenic lines were higher in comparison to control (1.74). Line D6 showed highest water use efficiency (3.9 μM CO2m-2s-1) followed by D7 (3.55), D8 (3.39), D5 (3.22), D3 (3.11), D2 (2.91), D1 (2.85) and D4 (2.62) in the decreasing order (Figure 9).
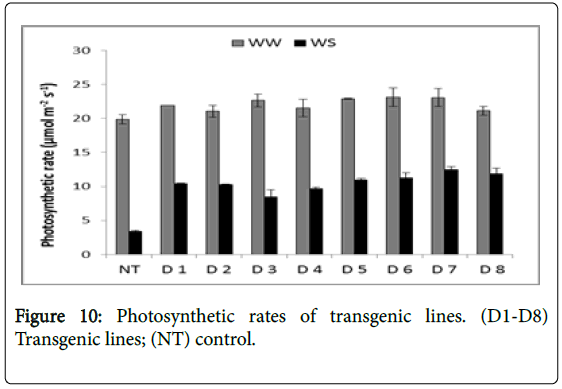
Photosynthetic rates ranged from 19.85 to 23.1 μM CO2 m-2s-1 under well watered condition but decreased to 3.37 to 12.45 μM CO2m-2s-1 under stress. Photosynthetic rates in transgenic lines were more compared to the control (3.37 μM CO2m-2s-1) under stress condition. Photosynthetic rates of the transgenic lines varied in the following decreasing order- D7 (12.45), D8 (11.83), D6 (11.21), D5 (10.94), D1 (10.39), D2 (10.27), D4 (9.67) and D3 (8.47) (Figure 10).
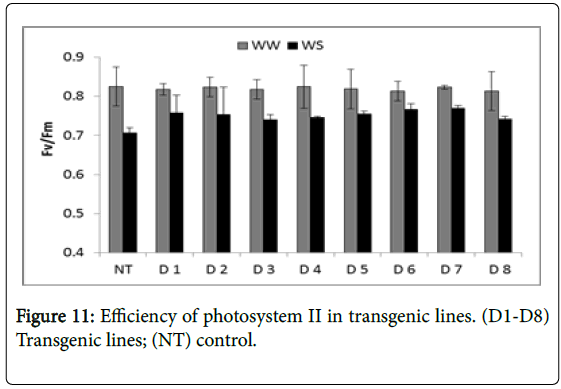
The photosynthetic capacity and the organization of the Photo- System II (PSII) reaction centre in the dark adapted leaves are associated. Higher organization of the photosystem II as represented by Fv/Fm ratio was found to be almost similar in transgenic and nontransgenic lines with slight increase in case of transgenic lines compared to non-transgenic lines. It ranged from 0.7 (control) to 0.76 (D7) respectively under water stressed conditions compared to 0.82 (control) and 0.82 (D7) respectively under well-watered conditions (Figure 11).
This result suggests that the higher efficiency of PSII reaction centre of the transgenic lines contributed to the increased photosynthetic capacity. This result suggested that increased photosynthetic capacity of the transgenic lines is due to the higher efficiency of PSII reaction centre.
Discussion
Drought is a frequent and periodic phenomenon occurring mainly in the drier months with varied intensities and is a major abiotic stress affecting about one fourth of the rice growing regions in Asia. About half of the total drought prone area in Asia belongs to Indian subcontinent. Surface water is evaporated due to increase in temperatures effecting the ground water levels and hydrologic cycle as a whole. Conventional breeding approaches were not able to address these challenges adequately as abiotic stress like drought is a polygenic trait. Transgenic approach provides a promising solution as the transfer of the gene of interest across the species is possible through this approach. In this study, an attempt was made to transfer the DREB1A gene from Arabidopsis thaliana into Pusa Basmati, an elite Indica rice cultivar, through Agrobacterium mediated transformation.
Various genes involved in dehydration tolerance are controlled by transcription factors and the enhanced expression of these transcription factors had resulted in improved drought tolerance [24]. One of the important aspects to be taken care is that there are varied levels of drought tolerance with respect to their constitutive or stress inducible expression of promoter genes. Marginal growth defects were found by the applications of constitutive promoters with DREB1A genes. Hence to address this problem in the present study we had utilized stress inducible RD29A promoter with AtDREB1A gene and effects on growth were not observed thus confirming the utility of RD29A promoter in transformation studies [25].
The molecular analyses i.e. PCR and Southern analysis could validate the incorporation of the foreign gene in the transgenic plants and the RT-PCR could successfully demonstrate the expression of the DREB1A gene in transgenic plants in comparison to controls (Figure 5). Decrease in leaf chlorophyll content is considered to be an indicative symptom during water deficit condition which results from oxidation and degradation of photosynthetic pigments, sustained photo-inhibition and photo breeding [26]. High chlorophyll content ensures continued photosynthesis under moisture stress enabling plant to resume its growth and development [27]. In our experiment, all the transgenic lines showed higher SPAD value (Table 1), compared to NT after 10 days of stress which is similar with the findings of Ravikumar et al. [28], but maintenance of high chlorophyll content under severe stress is imparted by osmotic adjustment which leads to higher leaf water content. Higher leaf water content maintains turgidity of cells reflecting continued growth, and metabolic activity in tissues hence leaf chlorophyll content is considered as an important trait for water stress tolerance [29]. High RWC ensures continued transpiration leading to canopy cooling effect and continued water uptake by roots. In transgenic plants, stomata closure has been observed which is considered as a prominent feature of increased dehydration tolerance [30]. Nayyar and Gupta [31] had reported that there is a decrease in relative water content in response to dehydration stress in different plants.
Leaves exhibit large reductions in RWC and water potential when they are subjected to drought. In our study, the transgenic plants maintained 74.3% of RWC (Table 1) at 11.05% of SMC compared to NT which has 53.2% of RWC at 11.36% of SMC. These results are in congruence with earlier reports stating that plants tolerate drought stress by maintaining high RWC during dehydration stress periods [32].
Moreover, maintenance of high chlorophyll content or high RWC is under the control of osmotic adjustment contributed by accumulation of different osmolytes including proline. Accumulation of proline may reduce stress-induced cellular acidification or prime oxidative respiration to provide energy needed for recovery. We observed that highest levels of proline were accumulated on 8th day of the stress in transgenic rice lines whereas the control plants had very low levels of proline during the same period [33].
Stomatal conductance increases under heat stress conditions and thereby evapotranspiration increases to keep the canopy temperature cool. This is reflected in our studies where the transgenic lines exhibited more stomatal conductance and transpiration rates compared to control. High light radiation and heat stress affect the electron transport chain. Photosystem II is highly heat sensitive and is affected during drought stress. Photo inhibition may occur during grain filling and this effect the rice yield. It was reported that drought stress reduces variable florescence (FV), and quantum yield [34]. Hence chlorophyll florescence measurement studies will help in understanding the equilibrium between metabolism and energy evolving processes which are affected during drought stress. Moreover chlorophyll florescence measurement is relatively simple technique which is non-destructive and less time consuming. Results in the present study show that there is a decrease in Fv/Fm ratio under stress conditions and this decrease is comparatively less in transgenic plants compared to control.
Transgenic lines and controls used in the study were given drought scores when subjected to stress as per IRRI, SES [35]. In this study, the controls (wild type) had a drought score 9 while transgenic lines D1, D3, D4, D6 and D8 had drought score 3 and the remaining transgenic lines D2, D5 and D7 showed a drought score 1. The recovery data suggests that all the transgenic lines showed recovery score of 1 (90% of all plants produce new leaves and tillers after irrigation applied 1-2 days), whereas the control showed a recovery score of 9 (less than 50% of all plants produce new leaves and tillers after irrigation applied for 7days) (Table 1).
Conclusion
From the results it is evident that the transgenic lines developed did show proper integration, expression and inheritance of the introduced gene. The transgenic lines when screened using different physiological parameters that are associated with drought tolerance, did express high levels of tolerance to drought under controlled conditions. AtDREB1A gene was also known to provide salt tolerance and studies are underway to test the view. Assessment of drought tolerance under field conditions where various stresses come into play is really challenging and at present, limitations do exist as biosafety issues are involved.
However, the results obtained are highly promising and transgenic lines developed can address drought tolerance in present day climate change scenario and could serve as valuable source material for future breeding activities.
Acknowledgements
This work was conducted with a funding grant from an Indian Council of Agriculture Research (ICAR)-Network Project on Transgenics in Crops (NPTC). Authors CKG and RSK are thankful to ICAR for awarding a Senior Research Fellowship to them. We are thankful to ICAR for providing AtDREB1A gene under the network project and to Director NRRI for all the facilities and support.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Pandey S, Bhandari H (2009) Drought, coping mechanisms and poverty-Insights from rainfed rice farming in Asia IRRI Discussion Paper Series No: 7-produced by the Asia and the Pacific Division, IFAD.
- Varshney RK, Bansal KC, Aggarwal PK, Datta SK Craufurd PQ (2011) Agricultural biotechnology for crop improvement in a variable climate: hope or hype? Trends Plant Sci 16: 363-371.
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsive-ness to drought, low-temperature, or high salt stress. Plant Cell 6: 251-264.
- Qin F, Sakuma Y, Li J, Liu Q, Li Y, et al. (2004) Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant Cell Physiol 45: 1042-1052.
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, et al. (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought, high-salt and cold responsive gene expression. Plant J 33: 751-763.
- Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, et al. (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47: 141-153.
- Xiao BZ, Chen X, Xiang CB, Tang N, Zhang QF, et al. (2009) Evaluation of seven function-known candidate genes for their effects on improving drought resistance of transgenic rice under field conditions. Mol Plant 2: 73-83.
- Chaitanya KG, Sai Krishna R, Mohan Dev T, GJN Rao (2013) Genetic variation in in vitro response of elite aromatic and non-aromatic rice varieties. Oryza 50:4 329-333.
- Vijayachandra K, Palanichelvam K, Veluthambi K (1995) Rice scutellum induces Agrobacterium tumefaciens vir genes and T-strand generation. Plant Mol Biol 29: 125-133.
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473-497.
- Saikrishna R, Chaitanya KG, Rao GJN (2013) Efficacy of different transformation methods in rice (Oryza sativa L.). J Experiment Biol Agri Sci 1: 106-110.
- Murray MG, Thompson WF (1980) Rapid isolation of higher molecular weight plant DNA. Nucl Acids Res 8: 4321-4325.
- Zoller MJ, Smith M (1982) Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucl Acid Res 10:Â 6487-6500.
- Sambrook J, Russell DW (2001) Molecular Cloning: A laboratory manual, 3rd ed, Cold Spring Harbor Laboratory Press, New York.
- Ramana Rao MV, Behera KS, Baisakh N, Datta SK, Rao GJN (2009) Transgenic indica rice cultivar ‘Swarna’ expressing a potatochymotrypsin inhibitor pin2 gene show enhanced levels of resistance to yellow stem borer. Plant Cell Tissue Organ Cult 99: 277-285.
- Liu AL, Zou J, Liu CF, Zhou XY, Zhang XW, et al. (2013) Over expression of OsHsfA7 enhanced salt and drought tolerance in transgenic rice. BMB Rep 46: 31-36.
- Bonnet M, Camares O, Veisseire P (2000) Effect of zinc and influence of Acremonium lolii on growth parameters, chlorophyll a fluorescence and antioxidant enzyme activities of rye grass (Lolium perenne L. cv Apollo). J Exp Bot 51: 945-953.
- Markwell J, Osterman JC, Mitchell JL (1995) Calibration of the Minolta SPAD-502 leaf chlorophyll meter. Photosynth Res 46: 467-472.
- Yadava UL (1985) A rapid and nondestructive method to determine chlorophyll in intact leaves. Hort Science 21: 1449-1450.
- Brady NC (1985) Advances in agronomy. Volume 38. Academic Press, New York, USA.
- Bates LS, Waldren RP, Teak TD (1973) Rapid determination of free proline for water stress. Plant Soil 39: 205-207.
- Nataraja KN, Jacob J (1999) Clonal differences in photosynthesis in Hevea brassiliensis. Mull Arg Photosynthetica 36: 89-98.
- Panda D, Sarkar RK (2013) Natural leaf senescence: probed by chlorophyll fluorescence, CO2 photosynthetic rate and antioxidant enzyme activities during grain filling in different rice cultivars. Physiol Mol Biol Plants 19: 43-51.
- Wang H, Wang H, Shao H, Tang X (2016) Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front Plant Sci 7: 67.
- de Paiva Rolla AA, de Fatima Correa, Carvalho J, Fuganti-Pagliarini R, Engels C, et al. (2014) Phenotyping soybean plants transformed with rd29A:AtDREB1A for drought tolerance in the greenhouse and field. Transgenic Res 23: 75–87.
- Farooq M, Wahid A, Kobayashi N, Fujita D, Basra SMA (2009) Plant drought stress: effects, mechanisms and management. Agron Sustain Dev 29: 185-212.
- Osakabe Y, Osakabe K, Shinozaki K, Tran LS (2014) Response of plants to water stress. Front Plant Sci 5: 86.
- Ravikumar G, Manimaran P, Voleti SR, Subhramanyam D, Sundaram RM, et al. (2014) Stress-inducible expression of AtDREB1A transcription factor greatly improves drought stress tolerance in transgenic indica rice. Transgenic Res 23: 421-439.
- Lawlor DW, Cornic G (2002) Photosynthetic carbon assimilation and associated metabolism in relation to water deficits in higher plants. Plant Cell Environ 25: 275-294.
- Datta K, Baisakh N, Ganguly M, Krishnan S, Shinozaki KY, et al. (2012) Overexpression of Arabidopsis and rice stress genes’ inducible transcription factor confers drought and salinity tolerance to rice. Plant Biotechnol J 10: 579-586.
- Nayyar H, Gupta D (2006) Differential sensitivity of C3 and C4 plants to water deficit stress: association with oxidative stress and antioxidants. Environ Exp Bot 58: 106-113.
- Moore JP, Vicre-Gibouin M, Farrant JM, Driouich A (2008) Adaptations of higher plant cell walls to water loss: drought vs desiccation. Physiol Plant 134: 237–245.
- Serraj R, Sinclair TR (2002) Osmolyte accumulation: can it really help increase crop yield under drought conditions? Plant Cell Environ 25: 333-341.
- Vazan S (2002) Effects of chlorophyll parameters and photosynthesis efficiency in difference beet. Assay Ph.D Islamic azad university science and research Tehran556) Branch, pp 285.
- International Rice Research Institute (IRRI) (1980) Standard Evaluation System for Rice, 2nd edition, International Rice Research Institute, Los Bafios, The Philippines, pp 44.
Citation: Geda CK, Repalli SK, Dash GK, Swain P, Rao GJN (2019) Enhancement of Drought Tolerance in Rice through Introgression of Arabidopsis DREB1A through Transgenic Approach. J Rice Res 7: 208.
Copyright: © 2019 Geda CK, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 3271
- [From(publication date): 0-2019 - Jan 31, 2025]
- Breakdown by view type
- HTML page views: 2607
- PDF downloads: 664