Research Article Open Access
Elimination of Antibiotic Multi-Resistant Salmonella Typhimurium from Swine Wastewater by Microalgae-Induced Antibacterial Mechanisms
Melissa Paola1, Jean Michel Prandinib1, Jalusa Deon Kich2 and Márcio Luís Busi da Silva3*
1Biotechnology and Sciences Program, West University of Santa Catarina, Videira, SC 89560-000, Brazil
2Department of Chemical Engineering, Federal University of Santa Catarina, Florianópolis, SC 88040-900, Brazil
3EMBRAPA Swine and Poultry, Concórdia, Brazil
- *Corresponding Author:
- Márcio Luís Busi da Silva
EMBRAPA Swine and Poultry
Concórdia, Brazil
Tel: +554934410456
E-mail: marcio.busi@embrapa.br
Received date: December 13, 2016; Accepted date: January 06, 2017; Published date: January 09, 2017
Citation: Paola M, Prandinib JM, Kich JD, Silva MLBd (2017) Elimination of Antibiotic Multi-Resistant Salmonella typhimurium from Swine Waste Water by Microalgae-Induced Antibacterial Mechanisms. J Bioremediat Biodegrad 8:379. doi: 10.4172/2155-6199.1000379
Copyright: © 2017 Paola M, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
The effect of microalgae-based swine waste water treatment on the removal of antibiotic multi-resistant Salmonella enterica serovar typhimurium was investigated. Photobioreactors (PBRs) containing diluted swine digestate with and without microalgae Scenedesmus spp. were inoculated with S. typhimurium (108 Colony Forming Units per milliliters - CFU mL-1). Viable cells of S. typhimurium were quantified over time by plate counts and qPCR amplification of the Salmonella invasion gene activator, hilA. In the absence of microalgae, S. typhimurium concentrations increased 1.5 log cells mL-1 in 96 h. In the presence of microalgae, S. typhimurium was completely eradicated within 48 h. In the PBRs with controlled pH (6.8 ± 0.8), concentration of S. typhimurium remained constant (2.8 ± 0.2 log CFU mL-1) throughout 96 h. Thus, natural increase in pH>10 due to photosynthesis was detrimental to the antibiotic multiresistant bacteria survival. Phycoremediation holds promises as an alternative for waste water treatment process for the elimination of the serious public health threatening antibiotic multi-resistant bacteria, thus effectively avoiding Salmonellosis outbreaks arising from animal farming activities.
Keywords
Antibiotic-resistant bacteria; hilA gene; Salmonella enterica serovar typhimurium; Scenedesmus spp.; Swine waste water
Introduction
Major concerns exist over the several invasive and antibiotic resistant organisms thriving in swine waste waters and that are known to threaten human and animal health. Among them, Salmonella enterica serovar typhimurium deserves special attention since it is the most prevalent antimicrobial resistant serovar in swines, and also frequently related to human infections and outbreaks [1].
Several physicochemical approaches are described to control the proliferation of pathogens, such as exposure to UV irradiation [2,3] use of strong oxidant radicals [2], pH increase [4], and selective membranes [5]. However, most if not all of these approaches are not economically feasible. Therefore, biological anaerobic digestion followed by stabilization ponds are the most common treatment option adopted in swine farming worldwide [3,6,7]. Pathogen elimination can occur under thermophilic conditions, but not under mesophilic conditions that prevail in most digesters [8].
Phycoremediation has been considered as and efficient tertiary treatment method to reduce organic compounds and nutrients from waste waters [9-12]. Some microalgae produce a wide variety of antibacterial substances that can inhibit or kill pathogens [13,14]. These waterborne pathogens may also be sensitive to high oxygen concentrations and increased pH produced by photosynthesis. Nevertheless, to the best of author’s knowledge, the mechanism in which phycoremediation reduce or even eliminate invase antibiotic multiresistant bacteria from swine waste waters has not been fully explored. This study demonstrates whether phycoremediation of swine waste waters could effectively control the proliferation of antibiotic-resistant Salmonella enterica serovar typhimurium. Ancillary objective include the evaluation of a pre-enrichment method for rapid quantification of S. typhimurium from complex environmental samples.
Materials and Methods
Photobioreactor conditions
Laboratory scale photobioreactors (PBRs) were used to simulate phycoremediation of swine waste water. Each PBR (3.5 L beakers) was filled with 3 L of diluted swine digestate (6%, v:v). Effluent physicochemical characteristics were (g L-1): pH 7.9, total solids (3- 8), total organic carbon (1.5-6.5), total inorganic carbon (0.8-1), total nitrogen (1.5-2), ammonia-N (0.9-1.5), phosphate-P (0.045-0.06). PBRs were subjected to mixotrophic conditions (12 h, light: dark) using red light emission diode light (PGL-RBC 2500, Parus) at 630 nm and 121.5 μmol m-2 s-1, room temperature (22 ± 2°C), and continuous mixing. PBRs were inoculated with Salmonella enterica Serovar typhimurium (105 CFU mL-1) and Scenedesmus spp. (30% v/v, 70 mg L-1 dry weight biomass), except the negative control without microalgae. To discern microalgae antibacterial effects from pH-derived photosynthesis, pH was adjusted daily in a set of PBRs by adding either 0.1 M HCl or 0.1 M NaOH. All experiments were conducted in duplicate.
Inocula source and identification
Microalgae was collected from a field scale facultative open pond used as tertiary treatment process downstream from a biodigestor at the Brazilian Agricultural Research Corporation (EMBRAPA) swine waste water facility (Concórdia, SC, Brazil). Scenedesmus spp. was the dominant microalgae in the PBRs as described elsewhere [9,10]. The strain was isolated and deposited in the collection of photosynthetic microorganisms for Agroenergy Research at Embrapa (Brasília, DF, Brazil) under access number Embrapa LBA#31 (IAN193.096).
Salmonella enterica serovar typhimurium was collected from a local river stream (Concórdia, SC, Brazil), characterized as a multi-resistant bacteria [1] and stored at the Embrapa’s microorganism bank collection under accession # 12301.
Analytical methods
Samples were analyzed twice a day for pH, temperature (pH– mV, Hanna Instruments, Inc.), dissolved oxygen (DO) (Lutron DO- 5519). Microalgae were quantified by optical density at 570 nm (Hach DR/2000). A satisfactory correlation (r2=0.97) between DW (gravimetric measuremets) and OD570 (mg-DW L-1=536.2 × OD570nm-36.89) was obtained. The specific growth rate μ (day-1) was calculated by fitting the microalgae dry weight obtained from samples within the first 24 hours of microalgae growth to an exponential function of (lnX-lnX0/t), where t was the time between the two measurements, and X and X0 (mg-DW L-1) were concentrations of biomass at t (24 hours) and t0, respectively. pH adjustments in all photobioreactors were performed with either 0.1 M HCl or 0.1 M NaOH.
S. typhimurium quantification – plate culture and qPCR
S. typhimurium inoculum was collected from an overnight culture plate, sub-cultured into nutrient broth (50 ml) and incubated in a shaker (100 rpm, 37°C, 24 h). Density of cell suspension was assessed with McFarland turbidity standard No. 05 (1-2 × 108 CFU mL-1). Samples from PBRs were diluted (100-10-5) in saline media (0.85%) and spread onto Chromogenic XTL4 agar plates (100 μL) to enumerate S. typhimurium colonies. Plates (duplicates from PBRs) were incubated overnight and results were reported as CFU mL-1.
Samples from PBRs were collected daily and diluted ten times in buffered peptone water for incubation at 37°C for 6 h – pre-enrichment step [15] prior DNA extraction (MoBio UltraClean Microbial DNA kit). Template genomic standard curves (10-1 to 10-10) were also subjected to the pre-enrichment step. hilA, a Salmonella gene required for pathogenicity invasion, was quantified by qPCR using specific primers [16]. Each 20 μL qPCR reaction mixture contained 2 × qPCRSYBR- Green mix (Ludwig Biotec, Brazil), 500 nM forward and reverse primers (Prodimol Biotecnologia®, Brazil), sterile DNAse-free water (Ludwig Biotec, Brazil) and 16 μg of DNA template. qPCR reactions were performed (7500 Applied Biosystems, The Netherlands) in a twostep thermal cycling procedure (95°C, 10 min; followed by 40 cycles of 95°C for 15 s and 62°C for 30 s). Quantitation of S. typhimurium was performed by interpolation from DNA template standard curves.
Statistical analysis
Tukey’s Pairwise Comparison test was used to determine significant difference between two data sets at 95% confidence level (p<0.05). All data were subjected to one-way analysis of variance (ANOVA) using OriginPro 8© OriginLab Corporation.
Results and Discussion
Scenedesmus spp. growth
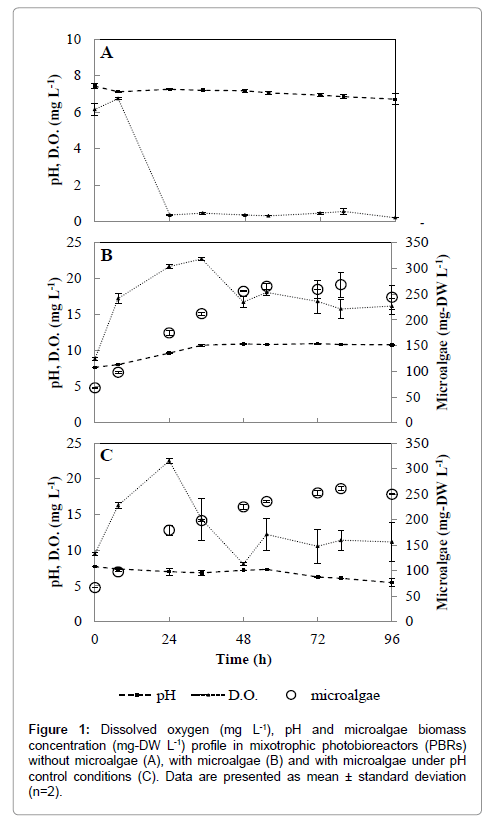
Scenedesmus spp. are known for their environmental ubiquity and predominance in waste stabilization ponds and high-rate algal ponds [17]. In the present study, microalgae growth rates of 96.5 ± 1.7 and 82.3 ± 3.4 mg-DW L-1 d-1 were reached in PBRs with and without pH adjustments, respectively. Specific growth rates (μ) were also calculated during the exponential growth phase of Scenedesmus spp. The mean value of μ in the interval from 0 to 24 h for PBRs with and without pH adjustments were 0.97 and 0.92 day-1, respectively. These values were within the higher growth rates reported for Scenedesmus sp. in swine waste waters [10,17]. The biomass increase measured in both PBRs treatments were very similar (p<0.05), with maximum microalgal concentration of 265 ± 3.9 mg-DW L-1 observed at 72 h (Figure 1). These experimental results are within typical values of maximal biomass production (0.2-1.0 g-DW L-1) reported for Scenedesmus spp. in domestic and swine waste waters from mixotrophic growth [17]. Interestingly, Scenedesmus spp. is a promising microalgal species that can thrive in waste waters with great performance on biomass production and lipid accumulation than other high-lipid-content microalgae [18]. Although not the scope of the present investigation, the obtained Scenedesmus spp. growth performance and biomass production are comparable to other studies focused on microalgal biomass to bioenergy conversion [19,20]. Thus, for a swine waste water treatment plant that uses tertiary phycoremediation treatment to remove pathogens and nutrients, microalgae biomass production could also aid in valuable biofuel products.
Phycoremediation effects on S. typhimurium
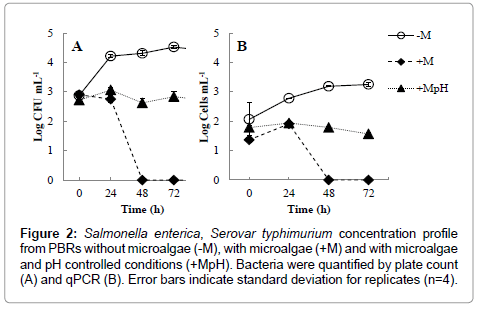
Salmonella spp. are able to thrive in different environments, survive several weeks in dry environments and several months in water, thus proliferation of S. typhimurium in swine waste water digestate was somewhat expected and later confirmed in the biological reactors without microalgae (Figure 2). In these reactors, S. typhimurium increased from 7.8 ± 1.9 × 102 CFU mL-1 (t=0 h) to a maximum of 3.4 ± 0.8 × 104 CFU mL-1 (t=72 h).
Figure 2: Salmonella enterica, Serovar typhimurium concentration profile from PBRs without microalgae (-M), with microalgae (+M) and with microalgae and pH controlled conditions (+MpH). Bacteria were quantified by plate count (A) and qPCR (B). Error bars indicate standard deviation for replicates (n=4).
Removal of pathogens from phycoremediation in ponds are usually attributed to a combination of long retention times, UV irradiation, high temperature, increased dissolved oxygen (DO) concentrations and high pH [21]. Among these factors, elevated DO and pH levels, resultant from optimal microalgal photosynthetic activities, have greater effects on pathogen inhibition and inactivation [22]. While it is possible to artificially increase DO and pH by aeration and chemicals addition, respectively, these practices are usually economically unfeasible and can be challenging to dosage at field scale. In this regard, phycoremediation could be alternatively considered as a disinfection step in waste water facility. S. typhimurium was completely removed from PBRs within 48 hours (Figure 2). In pH adjusted PBRs, S. typhimurium concentrations did not vary (p=0.12) throughout the entire experiment time frame and remained constant at 7.5 ± 1.4 × 102 CFU mL-1. Negligible difference in DO (p>0.05) concentrations were observed between PBRs treatments (Figure 1), thus discharging the probability of bacteria elimination by DO. The measured DO concentrations were above solubility levels (i.e., 8.3 mg L-1 at 1 atm and 25°C) (Figure 1). Oxygen saturation is normally found in ponds or photobioreactors due to high microalgae growth rate and associated photosynthetic activities [10,17,21]. The high pH values measured in the PBRs without pH adjustment seemed to play a major role on the inactivation of S. typhimurium during Scenedesmus spp. growth.
The antibacterial properties of microalgae exsudates should not be ruled out [14]. Scenedesmus sp. has been previously shown to act against Gram-positive (e.g., Staphylococcus aureus) and Gram-negative (e.g., Escherichia coli and Pseudomonas aeruginosa), but it was not very effective against Salmonella sp. [14,23]. This fact associated with the lack of S. typhimurium removal from the pH controlled PBRs suggests that microalgae metabolites had insignificant contribution on bacteria removal.
Sample preenrichment and qPCR fast approach
Colony count techniques consider CFU numbers, which can consist of more than one cell count and thus lead to data overestimation [24]. Therefore, to increase accuracy in bacterial concentration, qPCR quantification method was also performed in parallel. The plating method showed significantly higher cell concentrations (between 0.8 to 1.5 log differences) than qPCR (Figure 2). However, no differences (p>0.05) in S. typhimurium growth ratio (X/X0) over time was verified between these two bacterial quantification methods.
Conclusions
Phycoremediation of swine waste water digestate was very effective to produce valuable microalgae biomass and eliminate antibiotic multi-resistant S. typhimurium within 48 h. Pathogen removal was linked to the inhibitory effects of high pH (as high as ≅ 11) as a result of photosynthesis. Quantification of invasive S. typhimurium in environmental complex samples was demonstrated by preenrichment followed by qPCR. Phycoremediation of swine waste waters can be a promising treatment strategy to control the spread of antibiotic resistant bacteria in the environment. This can be particularly appealing when cogitating water reuse in animal farming and the risks associated with public health outbreaks from such practices.
Acknowledgements
The authors thank Remídio Vizzotto and Luiza Letícia Biesus from Embrapa Swine and Poultry for technical support. Authors thank financial support from Coordination for the Improvement of Higher Level or Education Personnel CAPESEMBRAPA (#001/2011) and the Brazilian Agricultural Research Corporation – EMBRAPA (#02.12.08.004.00.05).
References
- Palhares J, Kich J, Bessa M, Biesus L, Berno L, et al. (2014) Salmonella and antimicrobial resistance in an animal-based agriculture river system. Sci Total Environ 472: 654-661.
- Macauley J, Qiang Z, Adams C, Surampalli R, Mormile M (2006) Disinfection of swine wastewater using chlorine, ultraviolet light and ozone. Water Res 40:2017-2026.
- Jenkins M, Endale D, Fisher D, Adams M, Lowrance R, et al. (2012) Survival dynamics of fecal bacteria in ponds in agricultural watersheds of the Piedmont and Coastal Plain of Georgia. Water Res 46:176-186.
- Viancelli A, Kunz A, Fongaro G, Kich J, Barardi C, et al. (2015) Pathogen Inactivation and the Chemical Removal of Phosphorus from Swine Wastewater. Water, Air, Soil Pollut 226: 263-272.
- Masse L, Massé D, Pellerin Y (2007) The use of membranes for the treatment of manure: a critical literature review. Biosyst Eng 98: 371-380.
- Da Silva M, Cantao M, Mezzari M, Ma J, Nossa C (2015) Assessment of bacterial and archaeal community structure in swine wastewater treatment processes. Microb Ecol 70: 77-87.
- Hill V (2003) Prospects for pathogen reductions in livestock wastewaters: a review. Crit Rev Environ Sci Technol 33: 187-235.
- Metcalf E, Eddy H (2003) Wastewater engineering: treatment and reuse. 4th edn. McGraw Hill, New York, United States of America.
- Mezzari M, Da Silva M, Nicoloso R, Ibelli A, Bortoli M, et al. (2013) Assessment of N2O emission from a photobioreactor treating ammonia-rich swine wastewater digestate. Bioresour Technol 149: 327-332.
- Prandini J, Da Silva M, Mezzari M, Pirolli M, Michelon W, et al. (2016) Enhancement of nutrient removal from swine wastewater digestate coupled to biogas purification by microalgae Scenedesmus spp. Bioresour Technol 202: 67-75.
- Lowrey J, Brooks M, McGinn P (2014) Heterotrophic and mixotrophic cultivation of microalgae for biodiesel production in agricultural wastewaters and associated challenges. J Appl Phycol 27: 1-14.
- Rahman A, Ellis J (2012) Bioremediation of Domestic Wastewater and Production of Bioproducts from Microalgae Using Waste Stabilization Ponds. J Bioremediation Biodegrad 3: e113.
- Ghasemi Y, Moradian A, Mohagheghzadeh A, Shokravi S, Morowvat M (2007) Antifungal and antibacterial activity of the microalgae collected from paddy fields of Iran: Characterization of antimicrobial activity of Chroococcus dispersus. J Biol Sci 7: 904-910.
- Guedes A, Barbosa C, Amaro H, Pereira C, Malcata F (2011) Microalgal and cyanobacterial cell extracts for use as natural antibacterial additives against food pathogens. Int J Food Sci Technol 46: 862-870.
- Malorny B, Löfström C, Wagner M, Krämer N, Hoorfar J (2008) Enumeration of Salmonella bacteria in food and feed samples by real-time PCR for quantitative microbial risk assessment. Appl Environ Microbiol 74: 1299-1304.
- Brunelle B, Bearson S, Bearson B (2011) Salmonella enterica Serovar typhimurium DT104 invasion is not enhanced by sub-inhibitory concentrations of the antibiotic florfenicol. J Vet Sci Technol 2: 1-4.
- Wu Y, Hu H, Yu Y, Zhang T, Zhu S, et al. (2014) Microalgal species for sustainable biomass/lipid production using wastewater as resource: A review. Renew Sustain Energy Rev 33: 675-688.
- Xin L, Hong-ying H, Jia Y (2010) Lipid accumulation and nutrient removal properties of a newly isolated freshwater microalga, Scenedesmus sp. LX1, growing in secondary effluent. N Biotechnol 27: 59-63.
- Singh M, Reynolds D, Das K (2011) Microalgal system for treatment of effluent from poultry litter anaerobic digestion. Bioresour Technol 102: 10841-10848.
- Shen Q, Jiang J, Chen L, Cheng L, Xu X, et al. (2015) Effect of carbon source on biomass growth and nutrients removal of Scenedesmus obliquus for wastewater advanced treatment and lipid production. Bioresour Technol 190: 257-263.
- Butler E, Hung Y, Al Ahmad M, Yeh R, Liu R, et al. (2015) Oxidation pond for municipal wastewater treatment. Appl Water Sci pp: 1-21.
- Gupta S, Ansari F, Shriwastav A, Sahoo N, Rawat I, et al. (2015) Dual role of Chlorella sorokiniana and Scenedesmus obliquus for comprehensive wastewater treatment and biomass production for biofuels. J Clean Prod 115: 255-264.
- Ishaq A, Matias-Peralta H, Basri H, Muhammad M (2015) Antibacterial activity of freshwater microalga Scenedesmus sp. on foodborne pathogens Staphylococcus aureus and Salmonella sp. J Sci Technol 7: 858-871.
- Krämer N, Löfström C, Vigre H, Hoorfar J, Bunge C, et al. (2011) A novel strategy to obtain quantitative data for modelling: combined enrichment and real-time PCR for enumeration of salmonellae from pig carcasses. Int J Food Microbiol 145: S86-95.
--
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 4133
- [From(publication date):
January-2017 - Jul 11, 2025] - Breakdown by view type
- HTML page views : 3198
- PDF downloads : 935