Research Article Open Access
Electrochemical DNA Biosensor for Sequences Related to the Human Papillomavirus Type 16 using Methylene Blue
Elaine VM Souza1,3*, Gustavo A Nascimento1, Nataly Amorim de Santana1, Danielly S. Campos-Ferreira1, Juliana de Araújo Bibiano1, Mariana S Arruda1, Danyelly Bruneska1,2 and José L Lima-Filho1,2
1Laboratory of Immunopathology Keizo Asami (LIKA), Federal University of Pernambuco (UFPE), AV. Moraes s/n, 50670-901 Recife, PE, Brazil
2Department of Biochemistry, Federal University of Pernambuco (UFPE), AV. 12 Professor Moraes Rego s/n, 50670-901 Recife, PE, Brazil
3Nursing Department, Universidade Federal de Alagoas-UFAL, Arapiraca 15 Campus, AV. Manoel Severino Barbosa, s/n, Bom Sucesso-Arapiraca-AL, CEP:57309-005, Brazil
- Corresponding Author:
- Souza EVM
1 Laboratory of Immunopathology Keizo Asami (LIKA)
Federal University of Pernambuco (UFPE)
AV. Moraes s/n, 50670-901 Recife, PE, Brazil
Tel: +55 82 9931 0827
Fax: 82-3214 +55 1172
E-mail: elainevms@yahoo.com.br
Received Date: May 10, 2014; Accepted Date: July 28, 2014; Published Date: August 05, 2014
Citation: Souza EVM, Nascimento GA, Santana NA, Campos-Ferreira DS, Bibiano JA, et al. (2014) Electrochemical DNA Biosensor for Sequences Related to the Human Papillomavirus Type 16 using Methylene Blue. Biosens J 3:107. doi: 10.4172/2090-4967.1000107
Copyright: © 2014 Souza EVM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Biosensors Journal
Abstract
Several studies show that infections with high-risk human papillomaviruses (HPV), mainly HPV type 16, can lead to the development of tumors as the cervical cancer. A rapid and precise diagnosis of the precancerous lesions by HPV is extremely important for a successful treatment. The present paper describes an electrochemical DNA biosensor for specific sequence detection of the E1 HPV 16, using the methylene blue (MB) as indicator hybridization. In order to develop the sensor, a 21-mer ssDNA probe related to E1 HPV gene was immobilized on the work electrode (WE). The obtained optimum conditions for immobilization of HPVE1.16P on the activated WE was with an immobilization time of 5 min and probe concentration of 8 μM. The hybridization between the probe and its complementary sequence was studied by Differential Pulse Voltammetry (DPV). Some hybridization experiments with non-complementary oligonucleotides were carried out to study the selectively of the biosensor. The results showed that this DNA biosensor could be used for detection E1 HPV gene due to selectivity of the electrochemical measurement system. A detection limit of probe immobilized on electrode to its complementary sequence was 1.49 nM.
Keywords
HPV; DNA biosensor; Methylene blue
Introduction
The relationship between human papillomavirus (HPV) and the occurrence of tumors was first described in the cervix, and it is currently known that HPV is a necessary condition for the development of cancer in this region [1]. Virtually 100% of cervical cancers, the second commonest cancer in women worldwide, contain the HPV DNA sequences from a high-risk oncogenic genital HPV [2,3]. Several experimental evidences show that infections associated with HPV can lead to the development of tumors in a variety of sites as the anogenital region of male and female, breast, skin, nails, lungs and the head and neck [4,5].
The HPV can infect keratinocytes of skin and mucosa and it a double stranded DNA with approximately 8 kb [6]. Papilomaviruses are not classified by serotype but by genotype about 130 HPV types are knonw according to the sequence of the viral DNA [3] and new types are continually being found [7,8]. The HPV can be classified into high (e.g., HPV-16 and 18) or low-risk (e.g., HPV-11) [9], related with their potential to induce a cancer [10].
The early diagnostic of the presence of these viruses has been a very important tool to prevent a cancer development, and molecular diagnostic system has also improved within last few years. Molecular techniques are sensitive, reliable and effective [11], it is not yet possible to introduce these techniques in public health in developing countries because of their high cost. It is necessary to research and develop alternative methods for rapid and inexpensive detection of HPV in order to standardize the laboratory in public health for assisting the early diagnosis of cervical cancer [11].
Currently there has been much interest in developing electrochemical DNA biosensors for rapid and specific diagnosis of bacterial and viral infections, genetic diseases and among other applications [12-16]. All reports have revealed that the electrochemical biosensors has advantages such as speed, simplicity, low cost, high sensitivity, and relatively simple instrumental [17,18] and the possibility of real-time detection [19,20]. The DNA biosensors are effective in converting the event of hybridization of DNA in a signal that can be measured. DNA hybridization usually occurs between an oligonucleotide with a known sequence (probe) and an unknown complementary strand of nucleic acid from solution (target). The hybridization of the probe to the target sequence can be detected using appropriate hybridization indicators [21]. The hybridization indicator can bind DNA through reversible physical intercalation between base pairs or through electrostatic interaction [22]. Methylene blue (MB) is an aromatic heterocyclic that has been widely used as an electrochemical intercalator to monitor the DNA hybridization reaction, because MB binds specifically to guanine bases in ssDNA [23] and a lower current signal is observed following DNA hybridization. In this study, a simple method was proposed using MB to monitor the hybridization reaction. The electrochemical DNA biosensor based on the gene of HPV 16 was proposed. Due to the key role of HPV 16 in induction of cancer of the cervix, development low cost and small size device associated sensitive, specific and rapid detection methods will certainly provide important new tools for the diagnosis of disease.
Experimental Materials
Electrochemical experiments were performed using AUTOLAB PGSTAT 30 (METROHM AUTOLAB, Netherlands) and GPES 4.7 software package. The utilized two-electrode system was composed of a pencil graphite electrode (Total length 3 cm and 2.5 mm diameter) as the working electrode. In addition, reference electrode was screenprinted using Ag/AgCl ink (Electrodag-Acheson, USA) under gold wire and then dried at 60°C.
The experiments were performed in triplicate. All the synthetic oligonucleotides (targets and probes) were purchased from Integrated DNA Technologies (Belo Horizonte, Brazil) as lyophilized powder with the following sequences:
DNA probe_HPVE1.16P (21-mer sequence): GCAAAGGCAGCAATGTTAGCA
DNA target_HPVE1.16T (21-mer sequence): TGCTAACATTGCTGCCTTTGC
DNAnon-complementary (24-mersequence): CCAAAGCCTT TCAAAAAAAGATTTCCA
Mix sequence: (DNA targed_HPVE1.16R and DNA non-complementary).
The oligonucleotide stock solutions were prepared with 0.5 M acetate buffer (pH 5.0) and kept frozen. Tris base was obtained from Promega (USA), and sodium acetate was obtained from Sigma (USA). The UltraPure distilled Water was purchased from Invitrogen (USA), and reagents were of analytical grade.
Bioinformatics
The HPV 16 E1 nucleotides sequence was collected from the National Center for Biotechnology Information (NCBI) database, corresponding to GenBank_accession number NC_001526. Subsequently, HPV16 E1 was aligned using the software CLC Combined Workbench v 3.6.1 software. Target sequence (HPVE1.16T) and non-complementary sequences were also constructed using this software.
Preparation of the working electrode (WE)
The pencil graphite (type 4B, total length of 3 cm and diameter of 2.5 mm) was polished with emery impregnated disc to obtain a smooth surface. The body of the working electrode (WE) was coated well with silicone, resulting in a free area for immobilization of 28.50 mm2. The graphite electrode was washed with ultra-pure water to remove possible contaminants in the surface of the electrode. The polished surface of the working electrode was activated by applying a potential [24] of 1.8 V for 5 min in 0.5 M acetate buffer (pH 5.0).
Immobilization of the oligonucleotides probe
Immobilization of the HPVE1.16P (probe) was conducted following electrochemical pre-treatment of the WE.
The probe was immobilized on the activated WE by applying 0.5 V (24) within different period of time (1-10 minute) in 0.5 M acetate buffer solution (pH 5.0) containing oligonucleotides. The electrode was then rinsed with ultrapure water.
Hybridization
The hybridization was performed by immersing the probe-modified WE (WE_HPVE.16P) in a solution (50 μl) containing a known amount of the target oligonucleotides in acetate buffer (0.5 M) pH 5.0 with 300 rpm stirring. The hybridization was allowed to proceed for 15 minutes at 57°C. The electrodes were washed with Tris-HCl buffer (pH 7.0) for removing non-hybridized sequences. The same protocol was applied for the hybridization of probe with non-complementary sequences.
MB reduction measurements
The probe-modified WE was immersing into the 500 μM solution of MB for 10 minutes without applying any potential on the electrode. WE was rinsed with 20 mM Tris-HCl buffer (pH 7.0), MB reduction sign was measured at DPV. The same protocols were applied for probe modified WE following hybridization with complementary or noncomplementary sequences. All the experiments were performed in an electrode chemical cell.
Electrochemical analysis
The electrochemical analysis were carried out by Differential Pulse Voltammetry (DPV) (from -0.9 up to -0.1 V) in Tris-HCl buffer pH 7.0, with modulation amplitude of 50 mV and scan rate of 20 mV/s. The results were treated using the GPES software moving average baseline correction, using a “peak width” of 0.01.
Statistics analysis
Experimental data were analyzed by the software Statistica 8 using parametric tests. To evaluate statistical significance, the one-way ANOVA was used for the comparison of multi independent group data. A level of p<0.05 was assigned as significant.
Results and Discussion
Preliminary Investigation
The pencil graphite electrode is successfully applied as the working electrode [25,26]. Several studies demonstrate the utility of pencil electrodes and DNA probe for amperometric measurements of trace nucleic acids [13-15,27-29]. These transducers attract more attention than other methods due to their high sensitivity, small dimensions, low cost, and compatibility with microfabrication technology [28,30-32].
In the present work, we used this procedure for detection of oligonucleotides corresponding to E1 HPV 16 gene, in order to aid the diagnostic the HPV virus. The pretreatment of the carbon surface increases its roughness and hidrophilicity which is essential for DNA adsorption on the electrodes [14].
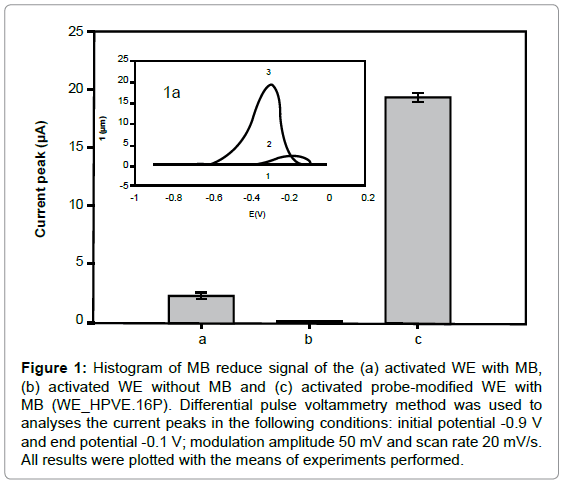
The activated WE without MB showed no significant current peak, while the activated WE with MB showed about 2.25 ± 0.21 μA (Figure 1). However, the MB voltammetric signal showed a higher peak with the WE_HPVE.16P (19.30 ± 0.39 μA), due to guanines free bases interaction of with MB. The increased was 84.44% when compared with activated WE with MB.
Figure 1: Histogram of MB reduce signal of the (a) activated WE with MB, (b) activated WE without MB and (c) activated probe-modified WE with MB (WE_HPVE.16P). Differential pulse voltammetry method was used to analyses the current peaks in the following conditions: initial potential -0.9 V and end potential -0.1 V; modulation amplitude 50 mV and scan rate 20 mV/s. All results were plotted with the means of experiments performed.
Effect of probe concentration
In the electrochemical DNA biosensor was immobilized E1 HPV 16 probe (WE_HPVE.16P) on the WE surface. The immobilization of the probe was with a potential 0.5 V in acetate buffer (0.5 M/pH 4.8) solution with increasing concentration of HPVE1 16P (1.5-20 μM). Previous studies demonstrate that this potential enhances stability of the immobilized probe, due to the electrostatic attraction between the positively carbon surface charged and the negative hydrophilic sugar-phosphate backbone charged with the bases oriented toward the solution ready to hybridize with the target [33].
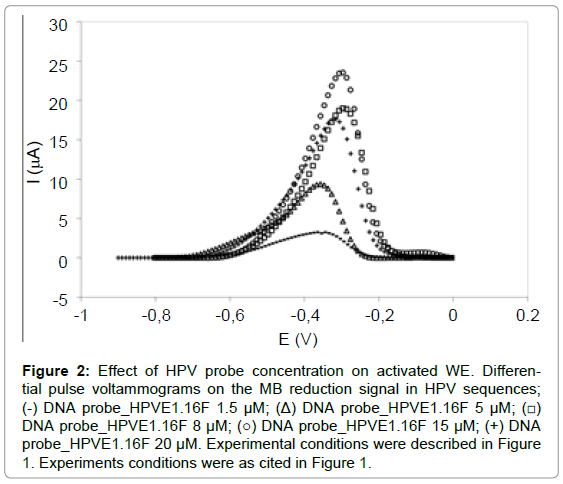
According to Figure 2, the concentrations 8 μM (19.3 ± 0.36 μA), 15 μM (21.9 ± 4.25 μA) and 20 μM (17.4 ± 1.95 μA) presented the highest signal and parametric statistical analysis (ANOVA-significant level p<0.05) demonstrated that the MB reduction signal was statistically similar. Therefore, concentration of 8 μM was suggested as good for the probe immobilization on the WE because of lower concentration and also presented better reproducibility than the concentration the 15 μM and 20 μM.
Figure 2: Effect of HPV probe concentration on activated WE. Differential pulse voltammograms on the MB reduction signal in HPV sequences; (-) DNA probe_HPVE1.16F 1.5 μM; (Δ) DNA probe_HPVE1.16F 5μM; (â�?¡) DNA probe_HPVE1.16F 8 μM; (â�?�?) DNA probe_HPVE1.16F 15 μM; (+) DNA probe_HPVE1.16F 20 μM. Experimental conditions were described in Figure 1. Experiments conditions were as cited in Figure 1.
Effect of immobilization time (probe)
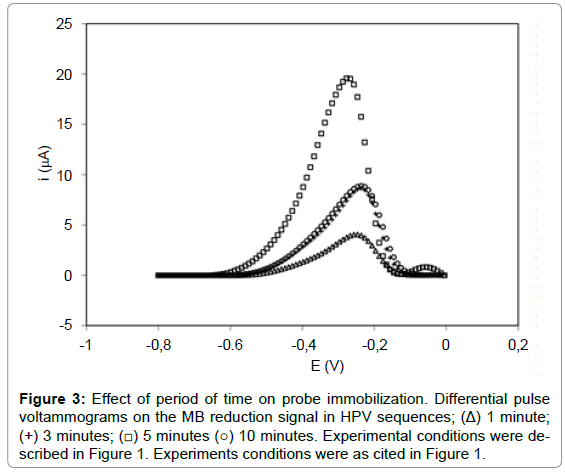
The probe immobilization time (HPVE1.16P) was investigated on WE in the 8 μM concentration at different times (1-10 min). According to Figure 3, 1 minute presented a current peak of 3.65 ± 0.40 μA, while the time of 3, 5 and 10 minutes showed respectively signal of 8.24 ± 0.22 μA, 19.30 ± 0.39 μA and 8.80 ± 0.53 μA, respectively. The results showed that the MB reduction signal increased with probe immobilization time up to 5 min (1-5 min), and then decreased significantly with 10 min (Figure 3). This decrease is attributed to the massive accumulation of the probe on the electrode surface, occurring probably overlapping probes, leading to a lower availability of guanine bases for MB interaction. Therefore, 5 min was suggested as optimum time for probe immobilization (8 μM). This immobilization time of 5 minutes is also found in a study of the human interleukine-2 in the construction of biosensors for the identification of recombinant plasmid with the gene sequence of interleukine-2 [28]. Results also were found in an electrochemical DNA biosensor with pencil graphite electrode for diagnosis of hepatitis B [31], detection of specific sequences of human papilloma virus (14), label-free electrochemical detection of the specific oligonucleotide sequence of dengue virus type 1 [15], electrochemical DNA biosensor for the detection of human papillomavirus E6 gene inserted in recombinant plasmid [34] and detection of Hepatitis C virus [35].
Figure 3: Effect of period of time on probe immobilization. Differential pulse voltammograms on the MB reduction signal in HPV sequences; (Δ) 1 minute; (+) 3 minutes; (â�?¡) 5 minutes (â�?�?) 10 minutes. Experimental conditions were described in Figure 1. Experiments conditions were as cited in Figure 1.
These results showed that probe concentration and the immobilization time affect the performance of the biosensor; therefore, these parameters are important for biosensor development.
Hybridization Detection
Detection of the hybridization was accomplished using reduction of the accumulated MB, that discrimination of the hybrid (DNADNA) from the probe (WE_HPVE.16P). The hybridization events were monitored by DPV utilizing 15 minutes with six different target concentrations (4, 7, 11 and 12 nM). MB reduction signal on the WE was around -0.3 V in 20 mM Tris-HCl buffer solution (pH 7.0). In this study, hybridization step was from the annealing with 300 rpm. The MB peak current from the probe-modified WE (19.3 ± 0.39 μA) before hybridization were bigger than the WE after hybridization in different concentrations. This happens due the interaction of the MB with single–stranded ssDNA by electrostatic binding [36].
The strong affinity of MB to the free guanine bases exposed to the exterior of ssDNA explains this decreased of the MB signal [37] in the double helix of the DNA hybrids. Thus, ssDNA and dsDNA identifies through the difference of the electrochemical signals. Several articles demonstrate that, electroactive indicators, as the MB, have frequently been used to characterize ssDNA and dsDNA by DPV [38].
The decrease in the MB signal is attributed to less MB accumulation on the DNA hybrid that causes the inaccessibility of MB to the guanine bases. It also is due to a steric inhibition of the reducible groups of MB packed between the bulky double helix of DNA hybrids [30].
After hybridization, the peak current response of MB reduction decreased around 13.5%-41.6%, according to the target concentration. However, a complementary DNA probe sequence electrostatically immobilized onto a glassy carbon electrode modified with chitosan for dengue using ferrocenium shows a difference of around 26%, before and after of the hybridization [39].
The experiment with non-complementary oligonucleotides was carried out to assess whether the suggested DNA sensor is selective via hybridization to the target. The current peak of MB observed at the WE_HPVE.16P after hybridization with non-complementary oligonucleotides was approximately equal the signal of the probe immobilized. The results demonstrated that the interaction between this sequence and immobilized probe did not lead to significant decrease in the MB reduction signal due to the absence of hybridization, being similar to the probe signal.
Electrochemical detection of HPV utilizing potential observed a slight decrease of the MB signal that is attributed to the small and irregular hybridization of non-complementary oligonucleotides with the probe sequences [14]. Irregular hybridization was not visualized in our study using in the hybridization process temperature.
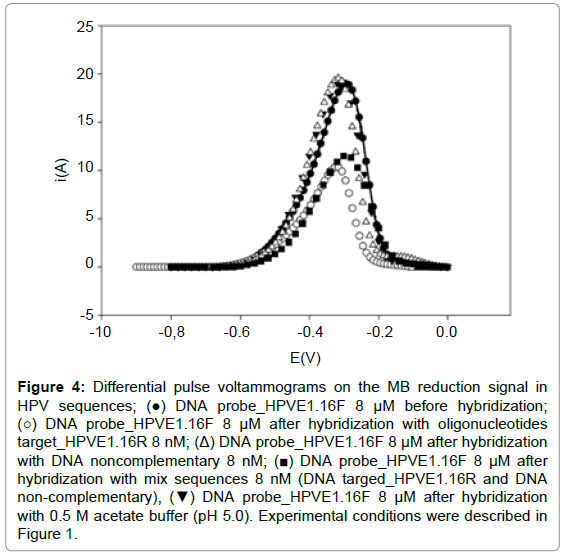
The experiments with mix of oligonucleotide sequence (complementary and non-complementary) demonstrated that the signal was equal to the WE after hybridization with complementary sequence (Figure 4). This is attributed to the process of hybridization of the probe with the HPVE1. 16T sequence present in the mix. These observations indicated that only complementary sequence could form a stable double strand DNA on the probe modified and thus demonstrating the selectivity of the biosensor proposed.
Figure 4: Differential pulse voltammograms on the MB reduction signal in HPV sequences; (â�?) DNA probe_HPVE1.16F 8 μM before hybridization; (â�?�?) DNA probe_HPVE1.16F 8 μM after hybridization with oligonucleotides target_HPVE1.16R 8 nM; (Δ) DNA probe_HPVE1.16F 8 μM after hybridization with DNA noncomplementary 8 nM; (â�? ) DNA probe_HPVE1.16F 8 μM after hybridization with mix sequences 8 nM (DNA targed_HPVE1.16R and DNA non-complementary), (â�?¼) DNA probe_HPVE1.16F 8 μM after hybridization with 0.5 M acetate buffer (pH 5.0). Experimental conditions were described in Figure 1.
The biosensor for detection of human interleukine-2 gene [30], dengue [15] and bovine papillomavirus [39] also present selectivity between complementary and non-complementary s39equences utilizing potential to hybridization
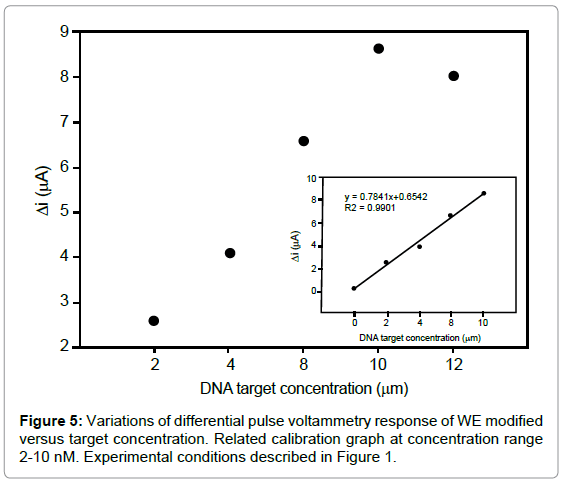
Quantitative analysis of target
The biosensor sensitivity was investigated by varying the concentration of the specific target, the difference between MB signals on the probe-modified WE in the absence and presence of target (ΔI) is increased with increasing the target concentration up to 10 nM (Figure 5). As seen in inset of figure 5, the calibration graph was linear between 2-10 nM with correlation coefficient of 0.9901. The regression equation was ΔI (μA)=0.7841x+0.6542, with a detection limit of 1.49 nM. The relative standard deviation over three independently modified probe electrodes measured at 8 nM of target was around 0.41% indicating a remarkable reproducibility of the detection method. Such detection limit compares favorably with those reported for the other electrochemical hybridization methods, such as, 5.78 nM detection of human interleukine-2 gene [30].
Conclusion
The effective immobilization of probe oligonucleotides related to HPV 16 on the electrode surface treaty was achieved. The optimum conditions for the construction of the working electrode were in the concentration of 8 μM and time of 5 minutes.
DNA biosensors utilizing graphite electrode for the detection of hybridization are useful for identifying specific nucleotide sequences (HPV) with selectivity. The hybridization of oligonucleotides on graphite was with annealing temperature using a MB reduction signal. The detection limit for this biosensor with complementary DNA target was of 1.49 nM within a hybridization time of 15 minutes.
New researches in biosensor techniques and hybridization methodology at the molecular level may contribute to future advancements in specific HPV diagnosis.
Acknowledgements
We are grateful for the financial support from the "National Council for Scientific and Technological Development (CNPq) - negligible Diseases 'and' Laboratory of Immunopathology Keizo Asami (LIKA).
References
- Bosch FX, Lorincz A, Muñoz N, Meijer CJL, Shah KV (2002) The causal relation between human papillomavirus and cervical cancer. JClin Pathol55:244-265.
- Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, et al (1999) Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 189:12–9.
- Stanley M (2010) Pathology and epidemiology of HPV infection in females. Gynecol Oncol117:S5-S10.
- ZurHausen H (2002) Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer2:342-350.
- Donne AJ, HampsonL, Homer JJ, Hampson IN (2010) The role of HPV type in Recurrent Respiratory Papillomatosis. Int J Pediatr Otorhinolaryngol Extra 74:7-14.
- Chen Z, SchiffmanM, Herrero R, Burk R (2007) Identification and characterization of two novel human papillomaviruses (HPVs) by overlapping PCR: HPV102 and HPV106. J Gen Virol88:2952-2955.
- Nobre RJ, Herraez-HernandezE, Fei JW, Langbein L, Kaden S, et al (2009). E7 oncoprotein of novel human papillomavirus type 108 lacking the E6 gene induces dysplasia in organotypic keratinocyte cultures. J Virol83:2907-16.
- Stanley M (2001) Human papillomavirus and cervical carcinogenesis. Best Pract Res ClinObstetGynaecol15:663-676.
- Campos MS, Roden RBS (2010) Papillomavirus Prophylactic Vaccines: Established Successes, New Approaches. J Virol 84:1214–1220.
- Fidelis AC, Dutra RF, Souza PRE, Melo COM (2013) A Simple HPV 18 Detection Method Based on Ultra Specific Primer Immobilized on Glass Slides. J Clin Lab Anal 27: 143–147.
- Mandong G, Yanqing L, Hongxia G, Xiaoqin W, Lifang F (2007) Electrochemical detection of short sequences related to the hepatitis B virus using MB on chitosan-modified CPE. Bioelectrochemistry 70:245–249.
- Dogan-Topal B, Uslu B, Ozkan SA (2009) Voltammetric studies on the HIV-1 inhibitory drug Efavirenz: the interaction between dsDNA and drug using electrochemical DNA biosensor and adsorptive stripping voltammetric determination on disposable pencil graphite electrode. BiosensBioelectron24:2358–2364.
- Sabzi RE, Sehatnia B, Pournaghi-Azar MH, Hejazi MS (2008) Electrochemical Detection of Human Papilloma Virus (HPV) Target DNA Using MB on Pencil Graphite Electrode. Journal of the Iranian Chemical Society 5:476-483.
- Souza E, Nascimento G, Santana N, Ferreira D, Lima M, et al. (2011) Electrochemical Detection of the Specific Oligonucleotide Sequence of Dengue Virus Type 1 on Pencil Graphite Electrodes. Sensors11: 5616-5629.
- Singh R, Mukherjee M, Sumana G, Gupta RK, Sood S, et al. (2014). Biosensors for pathogen detection: A smart approach towards clinical diagnosis. Sens Actuators B Chem 197:385-404
- Mono R, Stred M, Sturd E (2012) Application of Electrochemical Biosensors in Clinical. Diagnosis. J Clin Lab Anal 34: 22–34.
- Perumal V, Hashim U (2014) Advances in biosensors: principle, architecture and aplications. J Appl Biomed 12: 1-15.
- Palecek E, Fojta M, Tomschik M, Wang J (1998) Electrochemical biosensors for DNA hybridization and DNA damage. BiosensBioelectron13:621–28.
- Teles F, Fonseca L (2008) Trends in DNA biosensors. Talanta77:606-23.
- Ravan H, Kashanian S, Sanadgol N, Badoei-Dalfard A, Karami Z(2014) Strategies for optimizing DNA hibridazationon surfaces. Anal Biochem 444: 41-46.
- Millan KM,Mikkelsen SR (1993) Sequence-selective biosensor for DNA based on electroactive hybridization indicators. Anal Biochem65: 2317-2323.
- Yang IV, Ropp PA, Holden Thorp H (2002) Toward Electrochemical Resolution of Two Genes on One Electrode: Using 7-Deaza Analogues of Guanine and Adenine to Prepare PCR Products with Differential Redox Activity. Anal Biochem 74:347-354.
- Wang J, Rivas G, Fernandes JR, Paz JLL, Jiang M, et al. (1998). Indicator-free electrochemical DNA hybridization biosensor. Anal Chim Acta 375:197-203.
- Cheng C, Chang K-C, Chen C-S, Pijanowska DG (2011) Biosensor with Nano-gold Particle Modified Pencil Lead Carbon Electrode for Long-term Glucose Monitoring of Waste Tree Branch Hydrolysis. Journal of the Chinese Chemical Society58: 739-748.
- Teepoo S, Chumsaeng P, Nethan P, Prueprang W, Tumsae P (2012) Highly Sensitive Pencil-Based Renewable Biosensor for Hydrogen Peroxide Detection with a Novel Bionanomultilayer.Int. J. Electrochem. Sci 7: 4645-4656
- Mirmomtaz E, Ensafi AA, Soleimanian-Zad S (2009) Determination of amiloride using a ds-DNA-modified pencil graphite electrode based on guanine and adenine signals. Electrochim Acta 54:1141-1146
- Hejazi MS, Pournaghi-Azar MH, Alipour E, Karimi F (2008) Construction, electrochemically biosensing and discrimination of recombinant plasmid (pEThIL-2) on the basis of interleukine-2 DNA insert. BiosensBioelectron23:1588–94.
- Alipour E, Pournaghi-Azar MH, Parvizi M, Golabi SM, Hejazi MS (2011) Electrochemical detection and discrimination of single copy gene target DNA in non-amplified genomica DNA. ElectrochimActa56: 1925-1931.
- Pournaghi-Azar MH, Hejazi MS, Alipour E (2006) Developing an electrochemical deoxyribonucleic acid (DNA) biosensor on the basis of human interleukine-2 gene using an electroactive label. Anal Chim Acta570:144–150.
- Meric B, Kerman K, Ozkan D, Kara P, Erensoy S, et al. (2002) Electrochemical DNA biosensor for the detection of TT and Hepatitis B virus from PCR amplified real samples by using methylene blue. Talanta 56:837-846.
- Mohammadi LB, Klotzbücher T, Sigloch S, Welzel K, Göddel M, et al.(2014) In vivo evaluation of a chip based near infrared sensor for continuous glucose monitoring. Biosens Bioelectron 53:99-104.
- Pividori MI, Merkoçi A, Alegret S (2000) Electrochemicalgenosensordesing: immobilization of oligonucleotides onto transducer surfaces and detection methods. BiosensBioelectron 15:291-303.
- Campos-Ferreira DS, Souza EVM, Nsacimento GA (2014) Electrochemical DNA biosensor for the detection of TT and Hepatitis B virus from PCR amplified real samples by using methylene blue. Arab J ChemIn Press, Corrected Proof,Available online 11 June 2014.
- Pournaghi-Azar MH, Ahour F, Hejazi MS. (2009) Differential pulse voltammetric detection of Hepatitis C virus 1a oligonucleotide chain by a label-free electrochemical DNA hybridization biosensor using consensus sequence of Hepatitis C virus 1a probe on the pencil graphite electrode. Electroanalysis 21: 1822-1828.
- Kelley SO, Barton JK, Jackson NM, Hill MG (1997) Electrochemistry of methylene blue bound to a DNA-modified electrode. Bioconjug Chem 8:31-37.
- Zhu L, Zhao R, Wang K, Xiang H, Shang Z et al. (2008) Electrochemical Behaviors of Methylene Blue on DNA Modified Electrode and Its Application to the Detection of PCR Product from NOS Sequence. Sensors8:5649-5660.
- Lin X, Wu P, Chen W, Zhang Y, Xia X (2007) Electrochemical DNA biosensor for the detection of short DNA species of Chronic Myelogenous Leukemia by using methylene blue. Talanta72:468–471.
- Teles FRR, Prazeres DMF, Lima-Filho JL (2007) Electrochemical detection of a Dengue-related oligonucleotide sequence using ferrocenium as a hybridization indicator. Sensors 7:2510-18.
- Nascimento GAN, Souza EVM, Campos-Ferreira DS, Arruda MS, Castelletti CHM, et al. (2012) Electrochemical DNA biosensor for bovine papillomavirus detection using polymeric film on screen-printed electrode. BiosensBioelectron 38: 61-66.
Relevant Topics
- Amperometric Biosensors
- Biomedical Sensor
- Bioreceptors
- Biosensors Application
- Biosensors Companies and Market Analysis
- Biotransducer
- Chemical Sensors
- Colorimetric Biosensors
- DNA Biosensors
- Electrochemical Biosensors
- Glucose Biosensors
- Graphene Biosensors
- Imaging Sensors
- Microbial Biosensors
- Nucleic Acid Interactions
- Optical Biosensor
- Piezo Electric Sensor
- Potentiometric Biosensors
- Surface Attachment of the Biological Elements
- Surface Plasmon Resonance
- Transducers
Recommended Journals
Article Tools
Article Usage
- Total views: 16069
- [From(publication date):
December-2014 - Jul 13, 2025] - Breakdown by view type
- HTML page views : 11261
- PDF downloads : 4808