Eco-Friendly Biodegradation of Plastics by Intuitive Bacterial Strains from Polyethylene Polluted Sites and Optimizing its Culture Conditions by Response Surface Methodology (Rsm)
Received: 02-Jan-2023 / Manuscript No. jbrbd-23-86205 / Editor assigned: 05-Jan-2023 / PreQC No. jbrbd-23-86205 (PQ) / Reviewed: 19-Jan-2023 / QC No. Jbrbd-23-86205 / Revised: 23-Jan-2023 / Manuscript No. Jbrbd-23-86205 (R) / Accepted Date: 30-Jan-2023 / Published Date: 30-Jan-2023
Abstract
The aim of this study was to investigate the biodegradation of polyethylene by two bacterial strains isolated from polyethylene contaminated sites. This work revealed that the soil of areas contaminated with polyethylene is a good source of bacteria capable of decomposing plastic materials. The biodegradability of the two bacterial strains was evaluated by performing invasion studies, SEM analysis and Sturm assay. Strains capable of metabolizing complex plastics. The plastic samples tested in this study were plastic cups, plastic bags and milk carton lids. Among the samples, milk pods were shown to be optimally degraded (24.40%) by Pseudomonas sp. and plastic bags were found to have optimal degradability (20.40%) by Bacillus sp. after 45 days of incubation. In this study, the important variables affecting PE depletion of the contaminated sites by isolates B1 and B7 and the optimized values were determined by optimizing the values. Values were filtered using the central composite design (CCD) in the Response Surface Methodology.
Keywords
Biodegradation; Locations Contaminated With Polyethylene; Evolution of Co2; Ldpe; Sturm's Test; Plastic Weight Loss
Introduction
Approximately 140 million tons of synthetic polymers are produced each year worldwide [1, 2]. Because polymers are extremely stable, their degradation cycle in the biosphere is limited [3, 4]. Environmental pollution from synthetic polymers, such as plastic waste and water-soluble synthetic polymers in wastewater, has been recognized as a major problem [5, 6]. The use of certain microorganisms and enzymes to degrade polymers is classified as a method of polymer biodegradation (Premraj and Doble, 2005) [7, 8]. The word plastic comes from the Greek word "plastikos", which means "capable of being molded into different shapes" (Joel, 1995) [9, 10]. They are natural and synthetic macromolecules made up of smaller units called monomers linked together [11, 12, 13]. Examples of natural polymers include proteins, polysaccharides, and nucleic acids (Chandra and Rustgi, 1998) [14]. Synthetic polymers have been developed for their strength and resistance to all forms of degradation (Chiellini et al., 2003) [15, 16]. These and other characteristics, such as stiffness, permeability and transparency can be controlled by adjusting the polymer synthesis, molecular weight and/or by the use of substances. Specific additives [17, 18]. Plastic is defined as a polymer (solid material) which, when heated, becomes flexible and can be molded [19, 20]. They are castable nonmetallic compounds and materials made from them can be molded into any desired shape and size (Seymour, 1989). Plastic is commonly used for a variety of purposes including packaging, disposable diaper liners, agricultural films and fishing nets [21, 22]. Plastics and their use have become an integral part of every sector of the economy. Plastics are divided into two groups: thermoplastics and thermosetting plastics (Alauddin et al., 1995). Thermoplastics are plastics that do not undergo a chemical change in their composition when heated and can be molded over and over again while thermosetting resins are potentially prone to decomposition due to hydrolytic separation [23]. Of chemical bonds such as ester bonds or amide bonds (Muller et al., 2001) [24]. Thermosets can be melted and molded into different shapes. Plastic bags bring many benefits to human life but at the same time cause longterm harm. The collected waste includes all kinds, including plastic materials such as plastic bags, plastic cups, etc., when burned, will produce toxic gases that are dangerous to health. Inhaling these gases causes lung disease and cancer. The process of decomposing plastic bags takes about 1000 years. Biodegradable polymers are designed to break down when discarded by living organisms. Microbial degradation of plastics is due to enzymatic activities leading to the cleavage of polymer chains into monomers. Microorganisms use the polyethylene film as the sole carbon source, causing partial degradation of the plastic. They settle on the surface of the polyethylene film forming a biofilm. The hydrophobicity of the cell surface of these organisms has been shown to be an important factor in the formation of biofilms on the polyethylene surface, thereby increasing the biodegradability of the polymer. The purpose of this study was to isolate microorganisms from spilled soil and screen for microorganisms capable of degrading polyethylene and to identify microorganisms with a high potential to degrade plastic.
Materials and Methods
Collection of Soil Samples
Soil samples were collected from plastic landfill sites in Gudiyattam in sterile polythene bags and transported to the laboratory. Samples were treated with proliferation to improve the isolation of plasticdegrading microorganisms.
Enrichment of Plastic degrading bacterial strains
Growth of plastic degrading bacteria strain 1 g of plastic spilled from the soil sample was added to 100 ml of mineral salt medium (MSM) containing the plastic sample. The flask was kept in a shaking incubator (100 rpm) at room temperature for uniform distribution. After incubation, samples were successively diluted and inoculated on nutrient agar and incubated at room temperature for 24-48 h, eight morphologically different bacterial strains (B1-B8) were isolated.
Screening of Plastic Degrading Strains
The isolated strains were inoculated into a plastic sample supplemented with mineral brine medium as the sole carbon source and incubated in a rotary shaker at 37°C for 24 h. After incubation, the turbidity of the medium was examined for growth. Two superior isolates (B1 and B7) were selected for further studies based on their biomass yield (Inoue et al., 1991).
4.3(i) The characterization of strains were examined by the following methods and compared with textbooks on decisive bacteriology by Bergey (9th edition).
(i) Colony morphology
(ii) Microscopic examination
(iii) Biochemical characteristics
(i) Colony morphology
Plastic degrading strains isolated from soil samples were streaked aseptically in Nutrient agar plates and incubated at 37°C for 48 hours 48 h. After incubation, colony morphology was studied using standard microbiological criteria, with particular attention to pigmentation, diameter, colony elevation, margin, consistency, and opacity.
(ii) Microscopic examination
Gram staining is performed and cell morphology (Gram response, shape and arrangement of cells) is examined under the light microscope of the culture medium. Liquid grows exponentially.
(iii) Biochemical properties
Strains B1 and B7 were isolated according to various biochemical criteria such as IMVIC, Catalase, Oxidase, TSI, denitrification, Cetrimide resistance, Arginine dehydrolase, Lysine decarboxylase and sugar utilisation test.
Biodegradation test
The plastic samples used in this investigation were plastic cup (sample A), plastic bag 1 (sample B), plastic bag 2 (sample C), bottle cap (sample D) have different properties. These plastic samples were collected and transported to the laboratory to be tested for their biodegradability. Each plastic material was tested with strains B1 and B7.
Sample Pre-treatment
Selected plastic samples (A-D) were surface sterilized and UV irradiated (365 nm) according to the methods described by Johnson et al., (1992) and Carine et al., (2001) respectively. Films were sterilized with chemical Tween 80 and bleached (Lee et al., 1991).
Membrane culture and harvesting
Sterilized and disinfected (pre-weighed) resin samples were aseptically added to the sterilized basic mineral salt medium and were incubated at 125 rpm/ min for 24, h at 37°C, did not inoculate isolated cultures. After incubation, no clouding or granulation will ensure sterility. Then, the cultures were inoculated with pre-enriched bacterial strains B1 and B7. This experimental set-up was kept incubated at 125 rpm for one month at 37°C. Control was maintained for each type of plastic film in a grain less mineral salt medium. At the end of each week, plastic samples were collected, air-dried and weighed.
Biodegradation monitoring methods
Surface area changes and weight loss of plastic materials have been determined (Coma et al., 2006). The percentage weight loss of each plastic material used in this study was determined by the formula Material weight loss = Sample weight loss/Initial sample weight × 100.
CO2 evolution test– Modified Sturm Test
Storm test modified CO2 released after biodegradation of LDPE was determined in terms of weight and volume by the Sturm test (Muller et al., 1992).
(i) Preparation of LDPE powders
LDPE sheets are cut into pieces and soaked in xylene. It is then boiled for 15 min, during which time xylene will dissolve the LDPE film and the remainder is crushed with a strip glove. Powder LDPE obtained was washed with ethanol to remove xylene remaining and allowed to evaporate to remove ethanol. The powder was dried in a hot air oven at 60°C overnight (Shah, 2007)
(ii) Gravimetric analysis Test and control
Vials containing synthetic medium supplemented with LDPE powder were prepared. The bacterial strain was added to the test bottle. Pass sterile air through 1 M KOH solution containers (air pretreatment flasks). CO2-free air is introduced into the test cylinders. CO2 free air is used by inoculum releases CO2 (after converts LDPE) in absorbs bottles. The test was performed at room temperature for 1 week. After 1 week amount of CO2 produces out of absorbs bottles is calculates equals adds 0.1 M Bacl2 which forms a precipitates out of barium carbonate.
(iii) Volumetric Analysis
Dissolved carbon dioxide present in the medium is also volumetrically measured by titration. Place 25 ml of the sample in a conical flask, add 0.05 ml of 0.1 N Thiosulphate solutions. Then add 2 drops of methyl orange indicator and titrated with 0.02 M sodium hydroxide solution. The end point is the color change from red-orange to yellow. Then add two drops of phenolphthalein indicator and continue titration until pink color appears. The volume of the titrants used was recorded, and the amount of CO2 escaped of was calculated using the according to the formula:
CO Evolution = A X B X 50 X 1000 / V, where
A= ml of NaOH titration solution,
B= titration of NaOH,
V= ml of sample.
Analysis by scanning electron microscopy
Control and test plastic samples (sample D and sample B) with bacterial strains invading during 45 days were analyzed by scanning electron microscopy.
Experimental Design
The Response surface Method was used to study the effect of independent variables including temperature (X1), time (X2), pH (X3) and CO2 generation (X4) to Variation response of plastic-degrading bacteria (B1 and B7).
B1 Isolate – Pseudomonas sp
Response surface methodology was used to investigate the effect of independent variables, including Temperature (X1), Time (X2), pH (X3), and CO2 evolved (X4) on response variable Yield from the plastic degrading bacteria B1. RSM design along with coded and uncoded levels is presented in [Table 6]. Central composite design (Five levels) and quadratic model was used to design this experiment. 30 treatments including 8 axial points, 16 fractional factorial points and six central points were randomly performed according to CCD, which is summarised in [Table 7].
| Independent variables | symbol | -1 | 0 | 1 |
|---|---|---|---|---|
| Temperature | (℃) | 25 | 6 | 55 |
| Time | min | 3 | 5.5 | 12 |
| pH | ---- | 4.5 | 5.5 | 10.5 |
| CO2 evolved | g/l | 2.6 | 6 | 3.15 |
Table 6: Independent variables and their corresponding levels (B1 isolate).
| Independent Variables | Response 1 | |||||
|---|---|---|---|---|---|---|
| Run | A:CO2 evolved | B:Time | C:pH | D:Temperature | Yield of CO2 after Degradation of LDPE - B1 isolate | |
| g/l | min | ℃ | g | |||
| 1 | 2.6 | 9 | 7.5 | 55 | 98.7565 | |
| 2 | 3.15 | 6 | 6 | 45 | 97.2787 | |
| 3 | 2.05 | 6 | 6 | 25 | 97.5331 | |
| 4 | 2.6 | 9 | 4.5 | 35 | 98.7687 | |
| 5 | 3.15 | 12 | 9 | 45 | 99.6865 | |
| 6 | 3.15 | 6 | 9 | 25 | 96.0282 | |
| 7 | 2.05 | 12 | 9 | 45 | 101.487 | |
| 8 | 2.6 | 9 | 10.5 | 35 | 94.6779 | |
| 9 | 3.15 | 12 | 6 | 25 | 99.5422 | |
| 10 | 3.7 | 9 | 7.5 | 35 | 96.3289 | |
| 11 | 1.5 | 9 | 7.5 | 35 | 99.3573 | |
| 12 | 3.15 | 6 | 9 | 45 | 104.716 | |
| 13 | 2.05 | 6 | 9 | 25 | 107.441 | |
| 14 | 2.05 | 12 | 9 | 25 | 103.603 | |
| 15 | 2.05 | 12 | 6 | 25 | 107.94 | |
| 16 | 2.6 | 9 | 7.5 | 35 | 99.8808 | |
| 17 | 2.6 | 9 | 7.5 | 35 | 103.156 | |
| 18 | 2.05 | 12 | 6 | 45 | 100.673 | |
| 19 | 2.05 | 6 | 9 | 45 | 101.888 | |
| 20 | 2.6 | 9 | 7.5 | 35 | 89.8269 | |
| 21 | 2.05 | 6 | 6 | 45 | 103.548 | |
| 22 | 3.15 | 12 | 6 | 45 | 106.888 | |
| 23 | 3.15 | 12 | 9 | 25 | 102.895 | |
| 24 | 2.6 | 9 | 7.5 | 35 | 99.8109 | |
| 25 | 2.6 | 15 | 7.5 | 35 | 99.3305 | |
| 26 | 2.6 | 9 | 7.5 | 35 | 96.6213 | |
| 27 | 2.6 | 3 | 7.5 | 35 | 103.937 | |
| 28 | 2.6 | 9 | 7.5 | 35 | 92.8462 | |
| 29 | 2.6 | 9 | 7.5 | 15 | 99.7928 | |
| 30 | 3.15 | 6 | 6 | 25 | 105.229 | |
Table 7: Experimental design for yield with independent variables, experimental and predicted values of response.
B7 Isolate –Bacillus sp
Response surface methodology was used to investigate the effect of independent variables, including Temperature (X1), Time (X2), pH (X3), and CO2 evolved (X4) on response variable Yield from the plastic degrading bacteria B7. RSM design along with coded and uncoded levels is presented in [Table 8]. Central composite design (Five levels) and quadratic model was used to design this experiment. 30 treatments including 8 axial points, 16 fractional factorial points and 6 central points were randomly performed according to CCD, which is summarised in [Table 9].
| Independent variables | symbol | -1 | 0 | 1 |
|---|---|---|---|---|
| Temperature | (℃) | 25 | 6 | 55 |
| Time | min | 6 | 5.5 | 12 |
| pH | ---- | 4.5 | 5.5 | 10.5 |
| CO2 evolved | g/l | 1.96 | 6 | 2.58 |
Table 8: Independent variables and their corresponding levels (B7 Isolate).
| Independent variables | Response 1 | |||||
|---|---|---|---|---|---|---|
| Run | A:Total CO2 evolved | B:Time | C:pH | D:Temperature | Yield of CO2 after Degradation of LDPE - B7 isolate | |
| g/l | min | ℃ | g | |||
| 1 | 1.96 | 12 | 10 | 25 | 103.17 | |
| 2 | 2.89 | 9 | 7.25 | 40 | 106.417 | |
| 3 | 2.27 | 9 | 7.25 | 40 | 95.6644 | |
| 4 | 2.58 | 6 | 10 | 25 | 109.375 | |
| 5 | 1.96 | 6 | 4.5 | 25 | 106.005 | |
| 6 | 1.96 | 6 | 10 | 55 | 105.683 | |
| 7 | 2.27 | 9 | 7.25 | 40 | 98.3261 | |
| 8 | 2.27 | 9 | 7.25 | 40 | 99.2521 | |
| 9 | 2.58 | 12 | 4.5 | 25 | 104.896 | |
| 10 | 2.27 | 9 | 7.25 | 40 | 95.5747 | |
| 11 | 2.27 | 9 | 7.25 | 70 | 96.8508 | |
| 12 | 2.58 | 12 | 10 | 55 | 99.9174 | |
| 13 | 2.27 | 9 | 7.25 | 10 | 105.054 | |
| 14 | 1.65 | 9 | 7.25 | 40 | 94.9121 | |
| 15 | 2.58 | 6 | 4.5 | 55 | 102.195 | |
| 16 | 2.27 | 9 | 7.25 | 40 | 100.251 | |
| 17 | 2.27 | 9 | 12.75 | 40 | 96.68 | |
| 18 | 2.58 | 6 | 4.5 | 25 | 97.416 | |
| 19 | 1.96 | 12 | 10 | 55 | 96.2601 | |
| 20 | 2.58 | 6 | 10 | 55 | 93.7492 | |
| 21 | 2.27 | 3 | 7.25 | 40 | 102.169 | |
| 22 | 1.96 | 6 | 10 | 25 | 100.014 | |
| 23 | 2.27 | 9 | 7.25 | 40 | 97.2635 | |
| 24 | 1.96 | 12 | 4.5 | 55 | 100.63 | |
| 25 | 1.96 | 6 | 4.5 | 55 | 100.045 | |
| 26 | 2.58 | 12 | 10 | 25 | 87.6247 | |
| 27 | 2.58 | 12 | 4.5 | 55 | 99.7923 | |
| 28 | 2.27 | 9 | 1.75 | 40 | 102.371 | |
| 29 | 1.96 | 12 | 4.5 | 25 | 104.613 | |
| 30 | 2.27 | 15 | 7.25 | 40 | 103.206 | |
Table 9: Experimental design for yield with independent variables, experimental and predicted values of response.
Statistical analysis
Experimental data were statistically analysed using design expert software. Numerous statistical parameters (lack-of-fit, predicted and adjusted multiple correlation coefficient and coefficient of variation) of different polynomials models were compared to select the best fitting polynomial model. To understand the effect of parameters and yield, response plots were generated using design expert software. All these experiments were performed in triplicates.
Screening and identification of strains of plastic-degrading bacteria
Soil samples were taken from plastic waste disposal areas in and around Gudiyattam [Figure 6]. The collected samples were treated to isolate 8 bacterial strains (B1-B8). Among them, strains B1 and B7 [Figure 7, 8] with optimal biomass were selected for further studies. Based on the morphological, cultural and biochemical characteristics, the isolates were identified as Pseudomonas sp. and Bacillus sp. [Table 1].
| Test for identification | Pseudomonas sp. (B1) | Bacillus sp. (B7) |
|---|---|---|
| Gram staining | -ve | +ve |
| Motility test | +ve | -ve |
| Catalase test | +ve | +ve |
| Oxidase test | +ve | +ve |
| Glucose | -ve | +ve |
| Sucrose | -ve | +ve |
| Lactose | -ve | +ve |
| Mannitol | -ve | +ve |
| IMVIC | -, +,+,+/- | -, +,+,+ |
| Triple sugar iron agar test | Kd / K, no H2S & no gas | -ve (no H2S, A+) |
| Urease test | -ve | -ve |
| Nitrate Reduction test | -ve | +ve |
| Cetrimide resistance test | +ve | -ve |
| Arginine dihydrolase test | -ve | -ve |
| Lysine decarboxylase test | -ve | -ve |
Table 1: Identification and characterization of pseudomonas sp. and bacillus sp. isolated from soil sample.
isolated from soil sample.Plastic biodegradation test
Selected plastic samples (sample A-D) were tested for biodegradation in the presence of Pseudomonas sp. [Figure 9] and Bacillus sp. [Figure 10] in the MSM environment. The most relevant practical criteria for determining the degradation of plastics in contact with the ground include deterioration in tensile properties and loss in weight.
(i) Surface changes in plastic samples
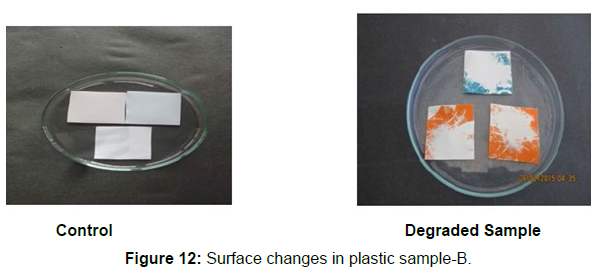
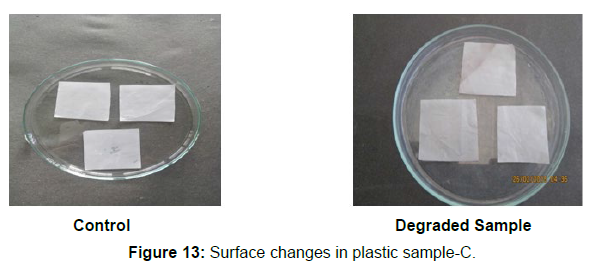
Different surface changes of plastic samples (samples A, B, C and D) incubated with resin-degradable soil isolates (B1 and B7) in MSM medium was observed [Figure 4, 5]. The surface of the plastic samples changed from smooth to rough with cracks indicating loss of tensile properties. The observed changes are shown in the figures [Figure 11, 12, 13, 14].
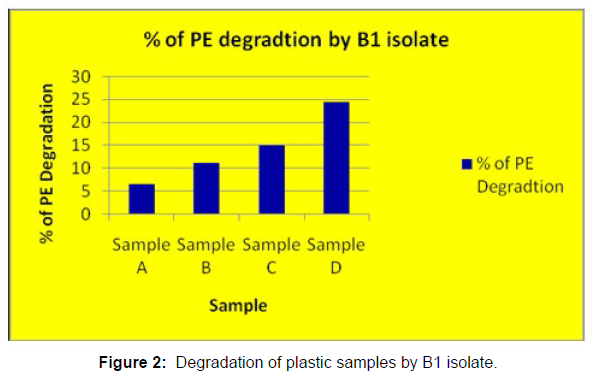
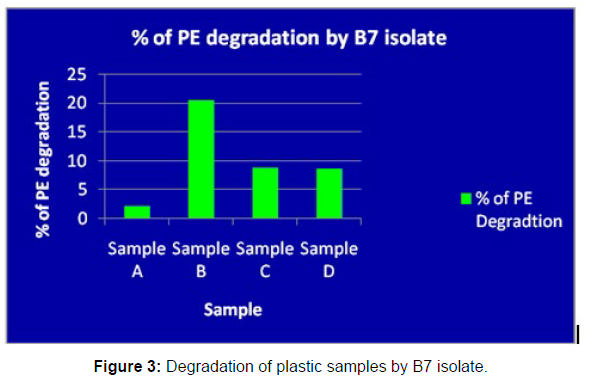
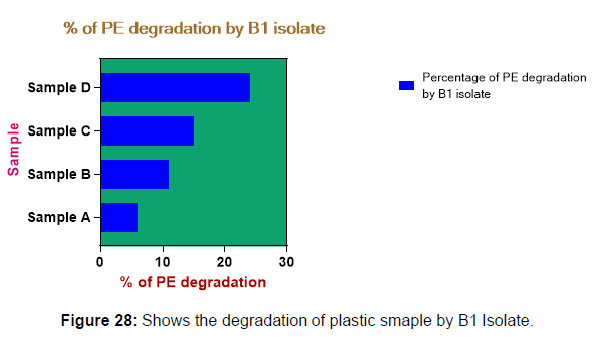
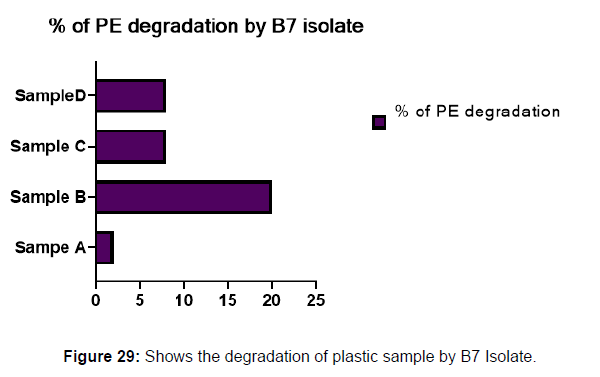
(ii) The obtained experimental results clearly demonstrated that Pseudomonas sp. (B1) and Bacillus sp. (B7) could mediate the degradation of all plastic samples tested in this study within 45 days. Among the different samples, sample D (milk skin) was shown to be optimally degraded (24.40%) by Pseudomonas sp. [Figure 2] and sample B (plastic bag 1) were found to be optimally degraded (20.40%) by Bacillus sp. [Figure 3]. [Tables 2] and [Table 3] show the weight loss of the plastic sample and its decomposition rate after 45 days of incubation.
| Samples | Weight of PE (g) | Weight of PE degraded (g) | Percent of PE degradation | |
|---|---|---|---|---|
| Initial | Final | |||
| Sample A | 0.091 | 0.085 | 0.006 | 6.50% |
| Sample B | 0.049 | 0.04 | 0.009 | 11.00% |
| Sample C | 0.045 | 0.041 | 0.007 | 15.00% |
| Sample D | 0.081 | 0.037 | 0.012 | 24.40% |
Table 2: Comparative analyses of the polyethylene (PE) weight before and after incubation for isolate B1.
| Samples | Weight of PE (g) | Weight of PE degraded (g) | Percent of PE degradation | |
|---|---|---|---|---|
| Initial | Final | |||
| Sample A | 0.091 | 0.089 | 0.006 | 2.19% |
| Sample B | 0.049 | 0.039 | 0.01 | 20.40% |
| Sample C | 0.045 | 0.041 | 0.004 | 8.88% |
| Sample D | 0.081 | 0.074 | 0.007 | 8.64% |
Table 3: Comparative analyses of the polyethylene (PE) weight before and after incubation for isolate B7.
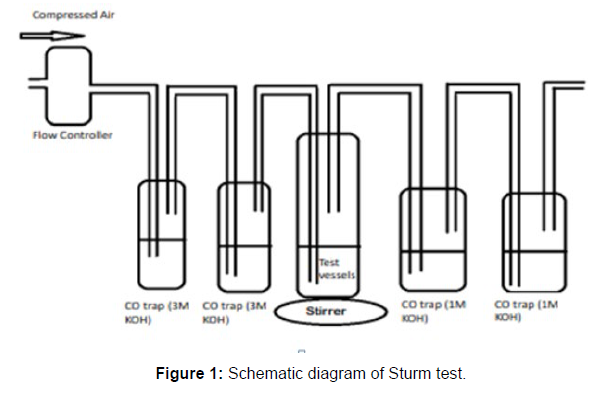
(ii) Sturm Test
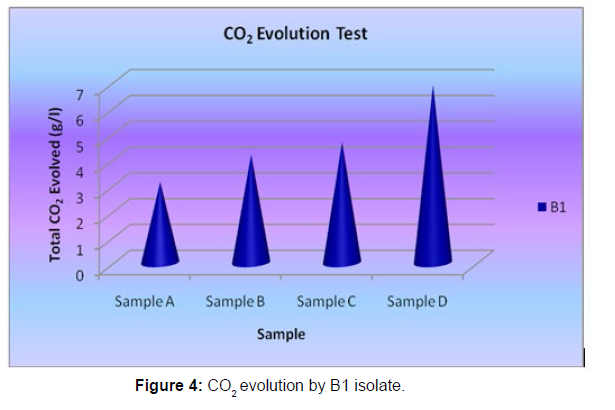
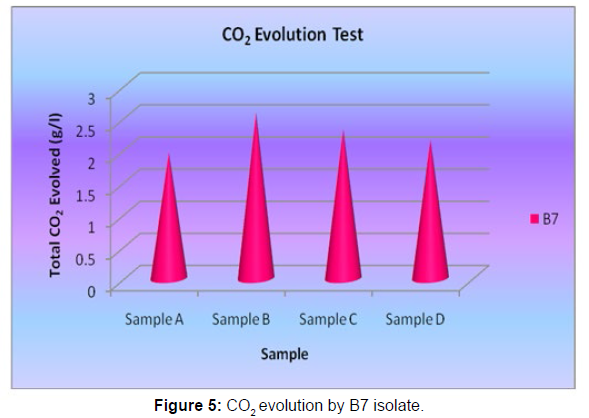
The Sturm test is commonly used to evaluate the biodegradability of polymeric materials [Figure 1]. Variable stability tests are used to measure CO2 in gaseous and dissolved forms in synthetic media [Figure 19]. The LDPE powder of the plastic samples was obtained by xylene treatment followed by grinding [Figure 15, 16, 17, 18]. The CO2 growth test provides valid data on the degradation rate of the plastic samples selected by Pseudomonas sp. [Figure 20] and by Bacillus sp. [Figure 21]. An optimal CO2 evolution (6.90 g/l) was observed from the treated sample D.
SEM Analysis
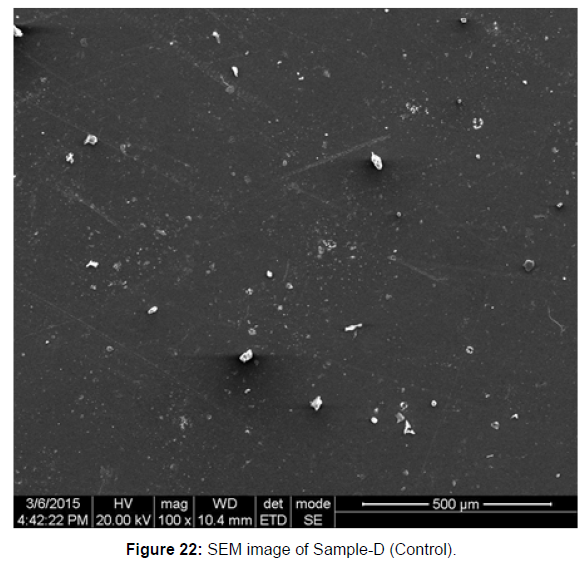
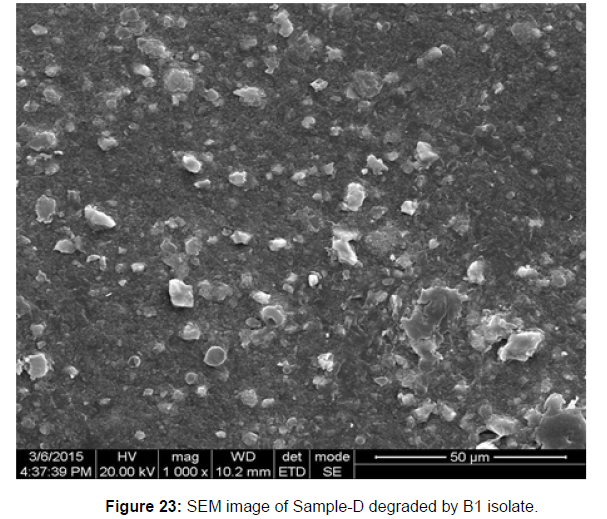
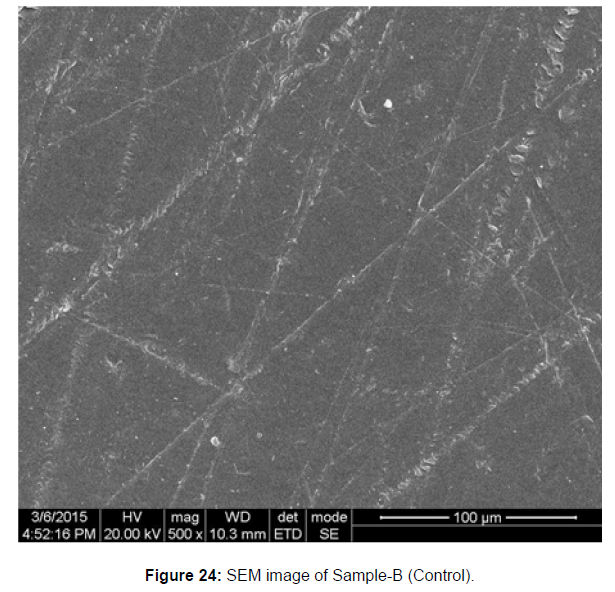
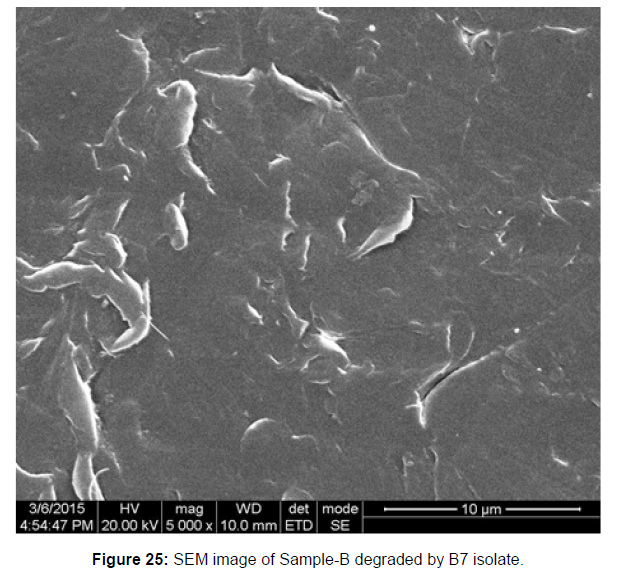
SEM image of the control plastic sample and the test samples (D and B) treated with Pseudomonas sp. (B1) and Bacillus sp. (B7) respectively showed major structural changes on their surface that was evident in our study [Figure 22, 23, 24, 25].
Discussion
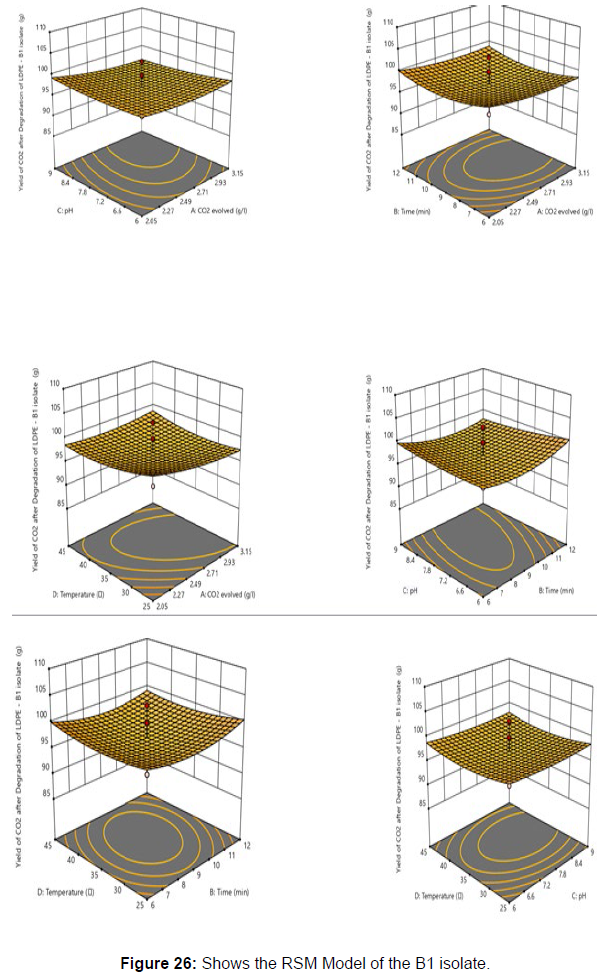
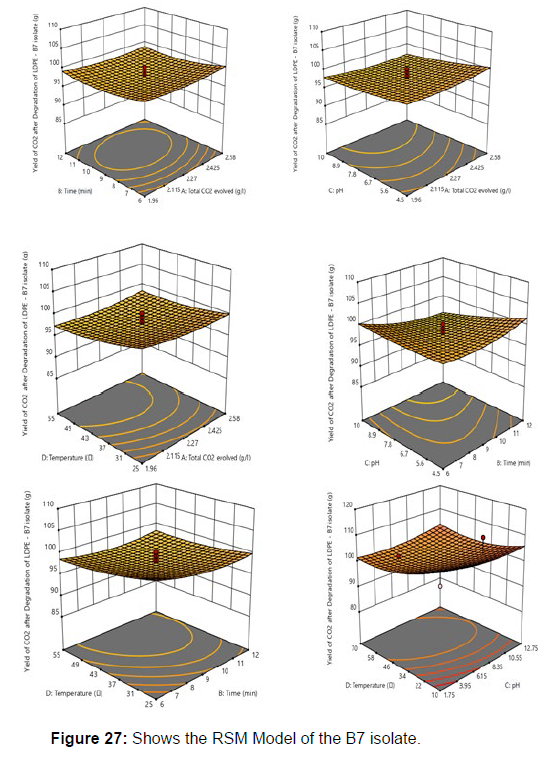
Plastics made from petroleum waste are called resins, which resist the degradation of dyes. In recent years, the public has become increasingly concerned about the environmental degradation associated with plastic disposal. Microorganisms such as bacteria and fungi are involved in the biodegradation of plastic. The present study focuses on the isolation, identification and degradation capabilities of soil plastic-degrading microorganisms [Table 4]. Two bacterial strains B1 and B7 have shown promising results for the biodegradation of plastics. Similarly, Aspergillus niger and Aspergillus japonicas showed degradation of low-density polyethylene after 30 days of incubation (Raaman et al., 2012). Pseudomonas putida isolated from garden soil samples showed its ability to degrade resins (Saminathan et al., 2014). Pseudomonas sp. and Bacillus sp. have shown their excellent tolerance to and decomposition of plastic materials [Table 5]. The surface of the plastic samples (D and B) changed from smooth to rough with cracks showing loss of tensile properties clearly indicating biodegradation of the resin [Figure 26, 27]. The hydrophobic nature of the plastic samples serves as a substrate for microorganisms to colonize their surfaces. This may be because extracellular compounds secreted by bacteria can disrupt the complex molecular structure of plastics (Priyanka and Archana, 2011). Among the different samples, sample D (milk skin) was found to be optimally degraded (24.40%) by Pseudomonas sp. and sample B (plastic bag 1) were found to be optimally degraded (20.40%) by Bacillus sp. after 45 days of incubation [Figure 28, 29]. The generation of CO during the Sturm test indicates a positive degradability test for polyethylene and this test fulfills the purpose of this study. The evolution of CO during the modified hardness test over a period of one week clearly shows positive results for the biodegradation of the plastic samples. From this present study, it can be concluded that both bacterial isolates can grow in minimal media with plastic as the sole carbon source. SEM images of the control and test samples (D and B) treated with Pseudomonas sp. (B1) and Bacillus sp. (B7) respectively showed large structural changes on their surface evident in our study. Similar results have been reported by Raaman et al., (2012). The biodegradation process is governed by many factors including the properties of the polymer, the type of organism and the nature of the pretreatment. Polymer properties such as mobility, crystallinity, molecular weight, functional groups and substituents present in its structure, as well as plasticizers or additives when added to the polymer all play an important role in polymerization. its decomposition. Therefore, further study of the microbial enzymes or organic acids involved in the degradation of polyethylene plastic will pave the way for the discovery of the exact degradation mechanism of the plastic material.
| Name of the isolate | Samples | Total CO2 evolved (g/l) |
|---|---|---|
| Pseudomonas sp. | Sample A | 3.14 |
| Pseudomonas sp. | Sample B | 4.2 |
| Pseudomonas sp. | Sample C | 4.68 |
| Pseudomonas sp. | Sample D | 6.9 |
Table 4: Quantification of CO2 evolution after degradation of LDPE ((1week) - B1
isolate.
| Name of the isolate | Samples | Total CO2 evovled (g/l) |
|---|---|---|
| Bacillus sp. | Sample A | 1.96 |
| Bacillus sp. | Sample B | 2.58 |
| Bacillus sp. | Sample C | 2.32 |
| Bacillus sp. | Sample D | 2.16 |
Table 5: Quantification of CO2 evolution after degradation of LDPE (1 week) - B7 isolate.
Conclusion
This study addressed the main concerns related to natural and synthetic polymers, their types, uses and biodegradability. Another area to be examined is the biodegradation of plastics by liquid culture, using bacterial strains. It is clear that most fastidious polymers can be degraded to some extent in the right environment at optimal concentrations. Microbial degradation of solid polymers such as polyethylene requires the formation of a biofilm on the polymer surface to enable bacteria to efficiently utilize insoluble substrates through enzymatic degrading activities. The growth of multicellular microbial communities known as biofilms, which attach to the surface of synthetic wastes, has been shown to be a potent degradation agent in nature. When considering the complete biodegradation of any organic substrate, the formation of a microbial population is essential to initiate the biodegradation process. Therefore, microbial colonization time is an important factor affecting the total decomposition time.
References
- Alauddin M, Choudkury IA, Baradie MA, Hashmi MSJ (1995) Plastics and their machining: a review. J Mater Process Technol 54: 40-46.
- Carine L, Adams T, Corinne VW, Christiane D (2001) The interaction mechanism between microorganisms and substrate in the biodegradation of polycaprolactone. J appl Polym Sci 83: 1334-1340.
- Chandra, RUSTGI, Renu Rustgi (1998) "Biodegradable polymers. Prog Polym Sci 7: 1273-1335.
- Chiellini E, Corti A, Imrie CT (2003) Biodegradation of thermally – oxidized, fragmented low density polyethylene. Polym Degrad Stab 81: 341-351.
- Coma C, Couturier Y, Pascat B, Bureau G, Gilbert S, et al. (2006 ) Estimation of the biodegradability of packaging materials by a screening test and a weight-loss method. Packag Technol Sci 7: 27-37.
- Inoue A, Yamamoto M, Horikoshi K (1991) Pseudomonas putida which can grow in the presence of Toluene. Appl Environ Microbiol 57: 1560-1562.
- Joel FR (1995) Polymer Science and Technology: Introduction to polymer science, Eds. 3. Pub: Prentice Hall PTR Inc., Upper Saddle River New Jersey 07458: 4-9.
- Johnson KE, Pometto AL, Nikolov ZL (1992) Degradation of degradable starch – polyethylene plastics in a compost environment. Appl Environ Microbiol 59: 1155-1161.
- Lee B, Pometto AL, Fratzke A, Bailey TB (1991) Biodegradation of degradable plastic polyethylene by Phanerochaete and Streptomyces species. Appl Environ Microbial 57: 678-685.
- Muller RJ (1992)"An interlaboratory investigation into biodegradation of plastics: Part 1: A modified Sturm-test." Mater Org 27: 179-189.
- Muller RJ, Kleeberg I, Dechwer WD (2001) Biodegradation of polyesters containing aromatic constituents. J Biotechnol 86: 87-95.
- Pooniah Saminathan, Anandhan Sripriya, Kaliappan Nalini, Thangavelu Sivakumar, Veerapandiyan Thangapandiyan, et al. (2014) Biodegradation of plastic by Pseudomonas putida isolated from garden soil samples. J adv botany zool 1: 2348-7313.
- Premraj R, Doble M (2005) Biodegradation of polymers. Indian J Biotechnol 4: 186-19
- Priyanka N, Archana T (2011) Biodegradability of Polyethylene and Plastic by the help of Microorganism: A Way for Brighter Future. Peng J Environ. Toxicol 1-4.
- Raaman N, Rajitha N, Jayshree A, Jegadeesh R (2012) Biodegradation of plastic by Aspergillus sp. isolated from polythene polluted sites around Chennai. J Acad Indus Res 1: 313-316.
- Seymour RB (1989) Polymer science before and after 1899: notable developments during the lifetime of Maurtis Dekker. J Macro mol Sci Chem 26: 1023-1032.
- Shah AA (2007) Role of microorganisms in biodegradation of plastics. 22: 51-53.
- Chakraborty P, Dey S, Parcha V, Bhattacharya SS, Ghosh A, et al. (2013) Design expert supported mathematical optimization and predictability study of buccoadhesive pharmaceutical wafers of loratadine. Biomed Res Int.
- Sawiphak S, Wongjiratthiti A (2021) Optimisation of culture conditions for pla-food-packaging degradation by bacillus sp. Snrusa4. Pertanika J Sci Technol 29: 407-425.
- Mehmood T, Ahmed A, Ahmad A, Ahmad MS, Sandhu MA, et al. (2018). Optimization of mixed surfactants-based β-carotene nanoemulsions using response surface methodology: An ultrasonic homogenization approach. Food Chem 179-184.
- Miloloža M, Ukić Š, Cvetnić M, Bolanča T, Kučić Grgić D, et al. (2022) Optimization of Polystyrene Biodegradation by Bacillus cereus and Pseudomonas alcaligenes Using Full Factorial Design. In Polymers.
- Meng K, Beng K, Mohd DS, Kassim AH, Razak A, et al. (2019) Optimization of Polystyrene Biodegradation using Response Surface Methodology (RSM) Measured by Simple Colorimetric Method. Int. J. Eng. Res. Technol 7: 216.
- Sahu R, Meghavarnam AK, Janakiraman S (2020) Response surface methodology: An effective optimization strategy for enhanced production of nitrile hydratase (NHase) by Rhodococcus rhodochrous (RS-6). Heliyon 6: e05111.
- Gajdhane SB, Bhagwat PK, Dandge PB (2016) Response surface methodology-based optimization of production media and purification of α-galactosidase in solid-state fermentation by Fusarium moniliforme NCIM 1099. Biotech 6: 1-14.
Indexed at, Crossref, Google Scholar
Indexed at, Crossref, Google scholar
Indexed at, Crossref, Google Scholar
Indexed at, Crossref, Google Scholar
Indexed at, Crossref, Google Scholar
Indexed at, Crossref, Google Scholar
Indexed at, Crossref, Google Scholar
Indexed at, Crossref, Google Scholar
Indexed at, Crossref, Google Scholar
Indexed at, Crossref, Google Scholar
Indexed at, Crossref, Google Scholar
Indexed at, Crossref, Google Scholar
Indexed at, Crossref, Google Scholar
Indexed at, Crossref, Google Scholar
Citation: Janarthanam H, Girirajan R, Balaji R, Prakasam J, Selvaraj V (2023) Eco-Friendly Biodegradation of Plastics by Intuitive Bacterial Strains from Polyethylene Polluted Sites and Optimizing its Culture Conditions by Response Surface Methodology (Rsm). J Bioremediat Biodegrad, 14: 549.
Copyright: © 2023 Janarthanam H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 3280
- [From(publication date): 0-2023 - Apr 03, 2025]
- Breakdown by view type
- HTML page views: 2826
- PDF downloads: 454