Dynamics of Higher-Order Chromatin Structure in Response to Viral Infection and its Potential for Treatment of Infectious Diseases
Received: 09-May-2022 / Manuscript No. JIDT-22-63211 / Editor assigned: 13-May-2022 / PreQC No. JIDT-22-63211 (PQ) / Reviewed: 27-May-2022 / QC No. JIDT-22-63211 / Revised: 01-Jun-2022 / Manuscript No. JIDT-22-63211 (R) / Published Date: 15-Jun-2022 DOI: 10.4172/2332-0877.1000506
Abstract
Viruses hijack cellular functions, primarily including the transcription machinery, through epigenetic mechanisms such as chromatin dynamics and Histone Post-Translational Modifications (HPTM). The spatial organization of chromatin is known to be highly dynamic in response to environmental stresses. In this review, we focus on our recent work demonstrating the dynamics of higher-order chromatin organization in response to influenza virus infection. We focused on the H4K20 trimethyltransferase Suv4-20h2, which interacts with cohesin in the uninfected condition. Upon viral infection, Suv4-20h2 binds to the viral protein and supresses its binding to cohesin. As a result, cohesin is loaded into a specific genomic region and a chromatin loop is formed in that region, resulting in gene expression that promotes viral replication. Inhibiting the binding of viral proteins to Suv4-20h2 is a potential therapeutic target for viral infections. In addition, H4K20me3 levels are rather decreased in non-infected patients with lung cancer and chronic lung disease, suggesting that H4K20me3 may be a biomarker for worsening viral infection. Thus, the dynamics of higher-order chromatin structures in response to viral infection may serve as a therapeutic target and biomarker for infectious diseases.
Keywords: Chromatin structure; Epigenetics; Histone modification; Infectious disease; Suv4-20
Introduction
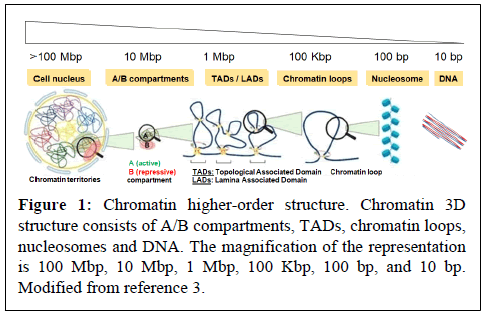
Virus hijacks cellular functions such as epigenetic transcriptional machineries, which are primarily regulated by chromatin dynamics, DNA methylation, Histone Post-Translational Modifications (HPTM) and non-coding RNA. In the nucleus of eukaryotic cells, chromatin is 3-Dimensionally (3D) organized into compartments (e.g. “active” A and “inactive” B compartment), Topologically Associated Domains (TADs) and loops, which control nuclear functions, including transcription (Figure 1) [1-3]. Individual TADs/loops are marked with active (e.g. H3K27ac, H3K4me3) or repressive (e.g. H4K20me3, H3K9me3, H3K27me3) histone modifications, which contribute to the epigenetic transcriptional regulation.
Accumulating evidence suggests that DNA boundary factors CTCF and cohesin are essential for the formation and maintenance of chromatin interaction domains [4-7]. The cohesin complex plays important roles in many aspects of chromosome biology, including not only sister chromatid binding, but also the regulation of transcription [8,9]. Cohesin is composed four core subunits, RAD21, SMC1, SMC3 and STAG proteins [9]. According to the loop extrusion model, cohesin plays a pivotal role in loop formation by loading into the CTCF binding site (CTCF-BS). This is regulated by factors that control cohesin loading (e.g. NIPBL and MAU2) and unloading (e.g. WAPL and its binding partner PDS5) [10-12]. Recent studies have suggested cohesin-mediated CTCF loop formation in various loci, including those of the immunoglobulin, β-globin, interferon gamma, IGF2-H19 and HOXA genes [7,13-19].
The 3D structure of chromatin is known to be highly dynamic, although it remains largely unknown how chromatin dynamics contributes to or modulates the infectious disease pathogenesis [20]. The RNA genome of influenza viruses does not directly integrate into the host chromatin, as it does for retroviruses or the Epstein–Barr virus [20,21]. In this review, we describe host chromatin dynamics and epigenetic regulation of influenza virus infection, with a particular focus on the role of the H4K20 trimethyltransferase Suv4-20h2.
Epigenetic Control in Influenza Virus Infection
Recent studies have revealed that Nonstructural protein 1 (NS1) of influenza A H3N2 subtype has a histone-like sequence and is modified by host histone modifying enzymes. The modified NS1 histone-like protein inhibited binding of the PAF1 transcription elongation complex to host histones, resulting in suppression of antiviral gene expression [22]. Histone Deacetylases 1 and 2 (HDAC1 and HDAC2) have been shown to be required for antiviral responses by the virus; expression of HDAC1 and HDAC2 is downregulated at both mRNA and polypeptide levels in influenza virus infection [23,24]. HDAC proteins control phosphorylation of Signal Transducer and Activator 1 (STAT1) and expression of interferon-stimulated genes (ISGs). Virus protein NS1 suppresses expression of interferon-stimulated genes through regulation of active H3K4me3 and repressive H3K27me3 levels [25]. The methylation state of H3K79 is elevated by influenza virus replication and regulates antiviral responses such as interferonmediated signaling [26]. In addition, viral NS1 also induces readthrough transcription and changes in chromatin structure; transcription elongation by RNA polymerase II has been shown to remodel the 3D genome structure during influenza virus infection [27].
Critical Role Of Suv4-20h2 In Influenza Virus Infection
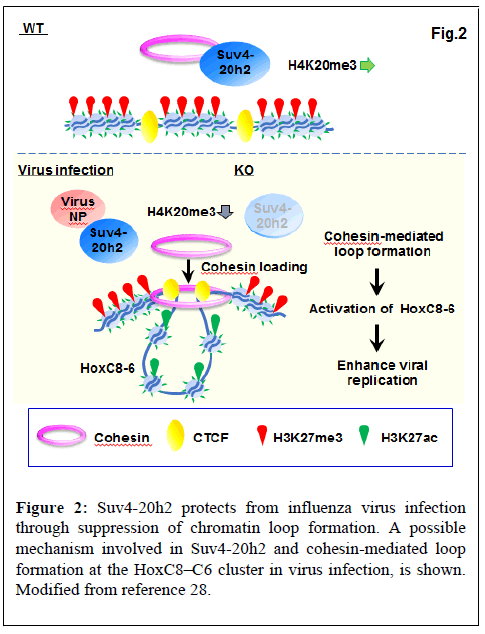
Recently, we have shown that the H4K20 trimethyltransferase Suv4-20h2 protects against influenza virus infection by regulating cohesin-mediated chromatin loop formation [28].
Suv4-20h2 supresses influenza virus replication
Suv4-20h1 and Suv4-20h2 are two homologous enzymes that methylate H4K20; Suv4-20h1 is responsible for dimethylation of H4K20 (H4K20me2) and Suv4-20h2 for trimethylation of H4K20 (H4K20me3) [29]. Loss of Suv4-20h2 is associated with loss of genomic integrity, increased senescence and telomere de-repression [30-32]. Suv4-20h2 was known to interact with cohesin [33]. H4-tail proteomics analysis demonstrated that H4K20me3 levels in lung tissues decreased over time following influenza viral infection, whereas H4K20me1 or H4K20me2 levels were unchanged [28]. We tested the effect of Suv4-20h deficiency on influenza viral infection using WT, Suv4-20h1 knockout (h1 KO), Suv4-20h2 KO (h2KO) and Suv4-20h1/Suv4-20h2 double-KO (dKO) Mouse Embryonic Fibroblasts (MEFs). H4K20me2 and H4K20me3 were lost in h1 KO and h2 KO/dKO cells, respectively [29]. h2KO and dKO cells showed higher viral replication compared to WT or h1 KO. These data suggest that loss of Suv4-20h2 promotes influenza virus replication in MEFs [28].
Influenza virus protein interacts with Suv4-20h2 and reduces its H4K20me3 activity
The Suv4-20h2 protein has two functional domains: the catalytic SET domain and the Clamp domain. The SET domain catalyzes H4K20me3, while the Clamp domain is responsible for binding to Heterochromatin Protein 1 (HP1) and recruitment of Suv4-20h2 to pericentric heterochromatin [33]. We showed that the SET domain of Suv4-20h2 interacts with influenza virus proteins (e.g. NP and PB2). In addition, Suv4-20h2, especially the SET domain, plays a role in inhibiting influenza virus replication. Furthermore, binding of viral NP proteins to the SET domain reduces the H4K20me3 activity of Suv4-20h2. Thus, upon infection, Suv4-20h2 binds to the viral protein, resulting in the inactivation of Suv4-20h2 [28].
Cohesin loads to the HoxC cluster upon influenza virus infection
Suv4-20h2 binds cohesin and has been shown to mediate chromocenter clustering [33]. In the uninfected condition, Suv4-20h2 interacts with heterochromatin-related proteins such as Rad21, Smc1, Smc3 (part of the cohesin multimer), Ezh2, CTCF, Nup98 and LaminB1. Viral infection suppressed the interaction of Suv4-20h2 with Rad21 and Smc1. Thus, cohesin is selectively detached from Suv4-20h2 by virus infection [28]. We found that influenza virus infection increased mRNA expression of transcription factors HoxC8, HoxC6, and HoxC5. Since Suv4-20h2 interacts with Ezh2, a core subunit of Polycomb Repressive Complex 2 (PRC2), we examined the binding profile of H3K27me3 that is a catalytic prodyct of Ezh2. H3K27me3 bound to the entire HoxC cluster in the uninfected WT cells, whereas binding was reduced at HoxC8-HoxC4 in virus-infected WT and dKO cells. In contrast, active H3K27ac bound to HoxC8- HoxC5 in virus-infected WT and dKO cells, but not in uninfected WT cells. For binding of H3K27me3 and H3K27ac, virus-infected WT cells showed a similar pattern as dKO cells. ChIP-seq also showed that bindings of CTCF and Rad21 are observed in the intergenic region between HoxC9 and HoxC8 (C9|8) and between HoxC6 and HoxC5 (C6|5). The results showed that inactivation of Suv4-20h2 by viral infection or gene deletion results in specific overlap of active (H3K27ac) and repressive (H3K27me3) histone markings on the HoxC cluster and exclusive activation in the C9|8 to C6|5 region [28].
As described above, cohesin proteins (Rad21 and Smc1) dissociated from Suv4-20h2 upon viral infection. ChIP-qPCR showed that in dKO and virus-infected WT cells, binding of Rad21 and Smc1 to the putative loop boundaries (C9|8 and C6|5) was increased. Thus, Suv4-20h2 most likely acts as an alternative cohesin unloading factor in a specific region (e.g. HoxC8-HoxC6). Inactivation of Suv4-20h2 by viral infection or gene deletion leads to active loop formation in HoxC8-HoxC6 through loading of cohesin into this region (Figure 2) [28].
HoxC8 and HoxC6 enhance viral replication through suppression of Wnt/β-catenin signalling
Knockdown of HoxC8 and HoxC6 significantly reduced viral replication [28]. Recent studies have shown that Wnt/β-catenin signaling is regulated by HoxC8 [34-36]. Consistent with this, genes in the Wnt/β-catenin pathway, such as secreted Frizzled-Related Protein 2 (sFRP2), were found to be highly expressed in Suv4-20 dKO MEFs [28]. sFRP2 is known to suppress Wnt signaling [37,38]. On the other hand, β-catenin inhibits influenza virus replication by promoting interferon-mediated responses [39]. β-catenin functions as a transcriptional regulator along with members of the T cell factor/ Lymphoid enhancer factor (Tcf/Lef1) family [40]. Recent studies have shown that the promoter region of Interferon regulatory factor 8 (Irf8) contains a consensus binding site for Tcf/Lef1, suggesting that Irf8 may be a target of Wnt/β-catenin signaling [41]. Active β-catenin accumulated in the nucleus following influenza viral infection in WT MEFs. In contrast, the nuclear accumulation of active β-catenin was reduced in the virus-infected dKO MEFs. Furthermore, treatment with recombinant sFRP2 protein suppressed nuclear transfer of active β- catenin and expression of Irf8 in virus-infected cells, while simultaneously increasing virus replication. Thus, HoxC8-HoxC6 promotes viral replication through sFRP2-mediated repression of β- catenin-Irf8 signaling [28].
Deletion of Suv4-20h2 exacerbates the pathology of influenza viral infection in mice
Deletion of Suv4-20h2 enhanced viral replication and exacerbated the pathology of influenza viral infection in vivo. Compared with WT, the h2 KO mice showed reduced survival, impairment of lung histology with severe pulmonary inflammation, and higher viral replication. Consistent with MEFs, compared with uninfected WT, h2 KO and infected-WT mice showed greater mRNA expression of HoxC8 and HoxC6 and lower Irf8 in lung tissues. Thus, HoxC8– HoxC6 mediated suppression of IFN signaling appears to enhanced viral replication in h2 KO mice [28].
Human lung cancer with lower Suv4-20h2/H4K20me3 enhances influenza viral replication
Recent studies have shown that influenza and Coronavirus Disease 2019 (COVID-19) are more severe and have higher mortality rates in patients with underlying diseases such as cancer [42-44]. Immunocompromised states due to cancer or its treatment are thought to be related to more severe pathology of virus infection. On the other hand, we and others demonstrated that H4K20me3 levels are reduced in breast cancer, colon cancer and lung cancer [45-47]. We have established an association between decreased Suv4-20h2/H4K20me3 and increased expression of HoxC6 and HoxC8. This has been reported for human cancer tissues, including prostate, cervical and esophageal cancers [48-51]. Our data showed that human lung cancer cells showing those changes are found enhanced viral replication, suggesting that Suv4-20h2 mediated chromatin dynamics might be a link to the process by which influenza virus infection becomes severe in patients with cancer [28].
Discussion and Conclusion
Suv4-20h2 binds to influenza virus proteins, and supresses binding to cohesion, which apparently triggers cohesin-mediated loop formation. In addition, repressive H3K27me3 and active H3K27ac marks show exclusive distribution, establishing selective expression at the HoxC locus. Thus, Suv4-20h2 is an alternative cohesin unloading factor at specific sites under conditions such as influenza virus infection. Furthermore, human cancer cells with reduced Suv4-20h2/ H4K20me3 levels and increased expression of HoxC6 and HoxC8 promote viral replication. This indicates that host epigenetic factors may indeed contribute to the severity of respiratory viral infections (e.g. influenza, COVID-19) in cancer patients. Collectively, the dynamics of higher-order chromatin structures in response to viral infection may serve as a therapeutic target and biomarker for infectious diseases.
Acknowledgment
We thank Prof. Susan Gasser and all members of our laboratory for helpful discussion.
Conflicts of Interest
The authors declared no conflicts of interest.
References
- Cremer T, Cremer C (2001) Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2:292-301.
[Crossref] [Google Scholar] [PubMed]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, et al. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376-380.
[Crossref] [Google Scholar] [PubMed]
- Alpsoy A, Sood S, Dykhuizen EC (2021) At the crossroad of gene regulation and genome organization: Potential roles for ATP-dependent chromatin remodelers in the regulation of CTCF-mediated 3D architecture. Biology (Basel) 10:272.
[Crossref] [Google Scholar] [PubMed]
- Zuin J, Dixon JR, van der Reijden MIJA, Ye Z, Kolovos P, et al. (2014) Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci USA 111:996-1001.
[Crossref] [Google Scholar] [PubMed]
- Sofueva S, Yaffe E, Chan WC, Georgopoulou D, Rudan MV, et al. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J 32:3119-3129.
[Crossref] [Google Scholar] [PubMed]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, et al. (2008) Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132:422-433.
[Crossref] [Google Scholar] [PubMed]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, et al. (2008) Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451(7180):796-801.
[Crossref] [Google Scholar] [PubMed]
- Guacci V, Koshland D, Strunnikov A (1997) A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91:47-57.
[Crossref] [Google Scholar] [PubMed]
- Wendt KS, Peters JM (2009) How cohesin and CTCF cooperate in regulating gene expression. Chromosome Res 17:201-214.
[Crossref] [Google Scholar] [PubMed]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, et al. (2017) Two independent modes of chromatin organization revealed by cohesin removal. Nature 551:51-56.
[Crossref] [Google Scholar] [PubMed]
- Busslinger GA, Stocsits RR, van der Lelij P, Axelsson E, Tedeschi A, et al. (2017) Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544:503-507.
[Crossref] [Google Scholar] [PubMed]
- Haarhuis JHI, van der Weide RH, Blomen VA, Yáñez-Cuna JO, Amendola M, et al. (2017) The Cohesin release factor WAPL restricts chromatin loop extension. Cell 169:693-707.
[Crossref] [Google Scholar] [PubMed]
- Degner SC, Wong TP, Jankevicius G, Feeney AJ (2009) Cutting edge: Developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol 182:44-48.
[Crossref] [Google Scholar] [PubMed]
- Hou C, Dale R, Dean A (2010) Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci USA 107:3651-3656.
[Crossref] [Google Scholar] [PubMed]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, et al. (2009) Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460:410-413.
[Crossref] [Google Scholar] [PubMed]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, et al. (2009) Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet 5:e1000739.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Xu M, Zhao GN, Liu GY, Hao DL, et al. (2015) Exploring CTCF and cohesin related chromatin architecture at HOXA gene cluster in primary human fibroblasts. Sci China Life Sci 58:860-866.
[Crossref] [Google Scholar] [PubMed]
- Watanabe T, Watanabe S, Kawaoka Y, et al. (2010) Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 7:427-439.
[Crossref] [Google Scholar] [PubMed]
- Dekker J, Belmont AS, Guttman M, Leshyk VO, Lis JT, et al. (2017) The 4D nucleome project. Nature 549:219-226.
[Crossref] [Google Scholar] [PubMed]
- Lesbats P, Engelman AN, Cherepanov P (2016) Retroviral DNA integration. Chem Rev 116:12730-12757.
[Crossref] [Google Scholar] [PubMed]
- West MJ (2017) Chromatin reorganisation in Epstein-Barr virus-infected cells and its role in cancer development. Curr Opin Virol 26:149-155.
[Crossref] [Google Scholar] [PubMed]
- Marazzi I, Ho JSY, Kim J, Manicassamy B, Dewell S, et al. (2012) Suppression of the antiviral response by an influenza histone mimic. Nature 483:428-433.
[Crossref] [Google Scholar] [PubMed]
- Nagesh PT, Husain M (2016) Influenza A virus dysregulates host histone deacetylase 1 that inhibits viral infection in lung epithelial cells. J Virol 90:4614-4625.
[Crossref] [Google Scholar] [PubMed]
- Nagesh PT, Hussain M, Galvin HD, Husain M (2017) Histone deacetylase 2 is a component of influenza A virus-induced host antiviral response. Front Microbiol 8:1315.
[Crossref] [Google Scholar] [PubMed]
- Menachery VD, Eisfeld AJ, Schäfer A, Josset L, Sims AC, et al. (2014) Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio 5:e01174-e01214.
[Crossref] [Google Scholar] [PubMed]
- Marcos-Villar L, Díaz-Colunga J, Sandoval J, Zamarreño N, Landeras-Bueno S, et al. (2018) Epigenetic control of influenza virus: Role of H3K79 methylation in interferon-induced antiviral response. Sci Rep 8:1230.
[Crossref] [Google Scholar] [PubMed]
- Heinz S, Texari L, Hayes MGB, Urbanowski M, Chang MW, et al. (2018) Transcription elongation can affect genome 3D structure. Cell 174:1522-1536.
[Crossref] [Google Scholar] [PubMed]
- Shiimori M, Ichida Y, Nukiwa R, Sakuma T, Abe H, et al. (2021) Suv4-20h2 protects against influenza virus infection by suppression of chromatin loop formation. iScience 24:102660.
[Crossref] [Google Scholar] [PubMed]
- Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, et al. (2008) A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev 22:2048-2061.
[Crossref] [Google Scholar] [PubMed]
- Jørgensen S, Schotta G, Sørensen CS (2013) Histone H4 lysine 20 methylation: Key player in epigenetic regulation of genomic integrity. Nucleic Acids Res 41:2797-2806.
[Crossref] [Google Scholar] [PubMed]
- Nelson DM, Jaber-Hijazi F, Cole JJ, Robertson NA, Pawlikowski JS, et al. (2016) Mapping H4K20me3 onto the chromatin landscape of senescent cells indicates a function in control of cell senescence and tumor suppression through preservation of genetic and epigenetic stability. Genome Biol 17:158.
[Crossref] [Google Scholar] [PubMed]
- Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, et al. (2007) Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J Cell Biol 178:925-936.
[Crossref] [Google Scholar] [PubMed]
- Hahn M, Dambacher S, Dulev S, Kuznetsova AY, Eck S, et al. (2013) Suv4-20h2 mediates chromatin compaction and is important for cohesin recruitment to heterochromatin. Genes Dev 27:859-872.
[Crossref] [Google Scholar] [PubMed]
- Yamamoto K, Chikaoka Y, Hayashi G, Sakamoto R, Yamamoto R, et al. (2015) Middle-down and chemical proteomic approaches to reveal histone H4 modification dynamics in cell cycle: Label-free semi-quantification of histone tail peptide modifications including phosphorylation and highly sensitive capture of histone PTM binding proteins using photo-reactive crosslinkers. Mass Spectrom (Tokyo) 4:A0039.
[Crossref] [Google Scholar] [PubMed]
- Lei H, Juan AH, Kim MS, Ruddle FH (2006) Identification of a Hoxc8-regulated transcriptional network in mouse embryo fibroblast cells. Proc Natl Acad Sci USA 103:10305-10309.
[Crossref] [Google Scholar] [PubMed]
- Lei H, Wang H, Juan AH, Ruddle FH (2005) The identification of Hoxc8 target genes. Proc Natl Acad Sci USA 102:2420-2424.
[Crossref] [Google Scholar] [PubMed]
- Klaus A, Birchmeier W (2008) Wnt signalling and its impact on development and cancer. Nat Rev Cancer 8:387-398.
[Crossref] [Google Scholar] [PubMed]
- Kaur A, Webster MR, Marchbank K, Behera R, Ndoye A, et al. (2016) sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. 532:250-254.
[Crossref] [Google Scholar] [PubMed]
- Hillesheim A, Nordhoff C, Boergeling Y, Ludwig S, Wixler V (2014) β-catenin promotes the type I IFN synthesis and the IFN-dependent signaling response but is suppressed by influenza A virus-induced RIG-I/NF-κB signalling. Cell Commun Signal 12:29.
[Crossref] [Google Scholar] [PubMed]
- Cadigan KM, Waterman ML (2012) TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol 4:a007906.
[Crossref] [Google Scholar] [PubMed]
- Scheller M, Schönheit J, Zimmermann K, Leser U, Rosenbauer F, et al. (2013) Cross talk between Wnt/β-catenin and Irf8 in leukemia progression and drug resistance. J Exp Med 210:2239-2256.
[Crossref] [Google Scholar] [PubMed]
- Hermann B, Lehners N, M Brodhun M, Boden K, Hochhaus A, et al. (2017) Influenza virus infections in patients with malignancies -- characteristics and outcome of the season 2014/15. A survey conducted by the Infectious Diseases Working Party (AGIHO) of the German Society of Haematology and Medical Oncology (DGHO). Eur J Clin Microbiol Infect Dis 36:565-573.
[Crossref] [Google Scholar] [PubMed]
- Kim YJ, Lee ES, Lee YS (2019) High mortality from viral pneumonia in patients with cancer. Infect Dis (Lond) 51:502-509.
[Crossref] [Google Scholar] [PubMed]
- Xia Y, Jin R, Zhao J, Li W, Shen H (2020) Risk of COVID-19 for patients with cancer. Lancet Oncol 21:e180.
[Crossref] [Google Scholar] [PubMed]
- Yokoyama Y, Matsumoto A, Hieda M, Shinchi Y, Ogihara E, et al. (2014) Loss of histone H4K20 trimethylation predicts poor prognosis in breast cancer and is associated with invasive activity. Breast Cancer Res 16:R66.
[Crossref] [Google Scholar] [PubMed]
- Özgür E, Keskin M, Yörüker EE, Holdenrieder S, Gezer U (2019) Plasma histone H4 and H4K20 trimethylation levels differ between colon cancer and precancerous polyps. In vivo 33:1653-1658.
[Crossref] [Google Scholar] [PubMed]
- Van Den Broeck A, Brambilla E, Moro-Sibilot D, Lantuejoul S, Brambilla C, et al. (2008) Loss of histone H4K20 trimethylation occurs in preneoplasia and influences prognosis of non-small cell lung cancer. Clin Cancer Res 14:7237-7245.
[Crossref] [Google Scholar] [PubMed]
- Miller GJ, Miller HL, van Bokhoven A, Lambert JR, Werahera PN, et al. (2003) Aberrant HOXC expression accompanies the malignant phenotype in human prostate. Cancer Res 63:5879-5888.
[Google Scholar] [PubMed]
- Alami Y, Castronovo V, Belotti D, Flagiello D, Clausse N (1999) HOXC5 and HOXC8 expression are selectively turned on in human cervical cancer cells compared to normal keratinocytes. Biochem Biophys Res Commun 257:738-745.
[Crossref] [Google Scholar] [PubMed]
- Du YB, Dong B, Shen LY, Yan WP, Dai L, et al. (2014) The survival predictive significance of HOXC6 and HOXC8 in esophageal squamous cell carcinoma. J Surg Res 188:442-450.
[Crossref] [Google Scholar] [PubMed]
- Narendra V, Rocha PP, An D, Raviram R, Skok JA, et al. (2015) CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347:1017-1021.
[Crossref] [Google Scholar] [PubMed]
Citation: Imai (2022) Dynamics of Higher-Order Chromatin Structure in Response to Viral Infection and its Potential for Treatment of Infectious Diseases. J Infect Dis Ther 10: 506. DOI: 10.4172/2332-0877.1000506
Copyright: © 2022 Imai Y. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 3228
- [From(publication date): 0-2022 - Apr 03, 2025]
- Breakdown by view type
- HTML page views: 2821
- PDF downloads: 407