Dynamic Changes in Aromatic Hydrocarbon Associated Catabolic Gene Profiles Linked to Aerobic and Anaerobic Microcosm Studies
Received: 27-Jan-2014 / Accepted Date: 28-Nov-2014 / Published Date: 01-Jan-2015 DOI: 10.4172/2090-5009.1000111
Abstract
The biodegradation of BTEX pollutants is often facilitated by both aerobic and anaerobic bacteria in the environment. Developing strategies to enable a rapid characterisation of related microbial populations is of importance to many industries, including the petrochemical industry. Here we use a qPCR assay to show that dynamic changes in key catabolic genes, from aerobic and facultative anaerobic bacteria involved in the breakdown of benzoate and toluene through microcosm studies, can be linked to the aerobic or anaerobic nature of these microcosms. This approach may prove useful in further studies of the biodegradation of pollutants by eubacteria in situ.
Keywords: qPCR; Bioremediation; Environment
17643Introduction
Aromatic compounds are one of the most prevalent types of compound present within our environment [1]. The BTEX family of aromatic compounds are of particular environmental importance due to their widespread occurrence, persistence in nature [2], potential to cause adverse human health effects, safety hazards, ecological and aesthetic impacts [3].
Aerobic pathways of aromatic hydrocarbon degradation employ oxygen as a terminal electron acceptor, and mono- and dioxygenase enzymes, to cleave the benzene ring which results in a loss of aromaticity [4]. Biodegradation pathways of aromatic hydrocarbons have been intensively investigated, much is known about the genes involved [2] and many related monooxygenase and dioxygenase genes have been characterised. Toluene dioxygenase, studied in detail by Gibson and colleagues [5] is an especially good illustration of a gene involved in the breakdown of one of the BTEX compounds. However, it is also the case that some microorganisms do not possess the toluene dioxygenase gene and instead transform toluene to benzoate, prior to aromatic ring cleavage by benzoate dioxygenase [6].
Under anaerobic conditions, BTEX compounds appear to be processed through the central benzoyl CoA reductase pathway-for a review readers are directed toward references [1] and [7]. Most BTEX degradation appears to be facilitated by facultative anaerobes, such as Thauera spp and Azoarcus spp [1] that functionalise the aromatic substrate via formation of a benzoyl CoA derivative before further ring degradation using reductive biochemistry [8].
Microcosms are widely used to both facilitate enrichment of pure cultures from environmental samples and allow studies of complex communities and their interactions with specific substrates to be performed. Previous studies have focused upon qualitative PCR approaches using catabolic genes as indicators of microbial degradation of compounds [9] or qPCR looking at one catabolic gene [10,11].
One key question that has not been extensively addressed in microcosm studies relates to the changes that occur in catabolic gene profiles in response to the addition of aromatic hydrocarbon substrates. If microcosms are to be used as a model for biodegradation in situ we need to provide evidence that such a dynamic change actually occurs.
As previously noted aerobic and anaerobic genes used by eubacteria for aromatic hydrocarbon degradation are very different. Therefore, the aim of this study was to monitor catabolic gene levels within both aerobic and anaerobic microcosms through qPCR. Any relationship linking gene profiles to the presence or absence of oxygen within degrading microbial communities could lead to direct inferences being made using a similar approach to microbial communities in environmental samples.
Materials And Methods
Microcosm construction
Benzoate degrading microcosms were established under both aerobic and anaerobic conditions using an inoculum of anaerobic river sediment (obtained from the River Lagan in Belfast, Northern Ireland) whilst toluene microcosms were set up using sediment from a petrochemical site contaminated with BTEX based in the UK. In the latter case, the inoculum was sediment from core material recovered at a depth of 6 metres below ground and stored in an air-tight container under nitrogen at 3°C prior to use.
Anaerobic microcosms were established inside a nitrogen glove box (Cole-Parmer). The box was flushed with nitrogen and media, vials, and soil were placed within the housing for at least 24 h prior to microcosm construction to allow the removal of dissolved oxygen. Inoculation of media, addition of resazurin and sealing of vials was carried out within the glove box.
Anaerobic and aerobic microcosms were constructed in 6 ml and 100 ml headspace vials (Supelco), respectively. 1 g of sediment was added to both microcosm types together with 4 ml of minimal salts media prepared as previously described [12]. Resazurin was added to the media at a concentration of 0.01 wt%, providing visual confirmation that all active anaerobic microcosms became anoxic within the first 24 h of incubation. Control microcosms were set up exactly as described above followed by autoclaving at 15 psi and 121°C for 15 min prior to incubation.
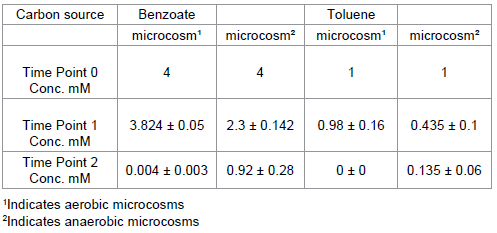
In total two experiments were constructed to monitor the degradation of benzoate (experiment 1) and toluene (experiment 2) under both aerobic and anaerobic conditions. In total 4 microcosm sets were constructed, in triplicate, comprising of :- (a) river sediment with benzoate as a sole carbon source, at a final concentration of 4 mM, incubated under aerobic conditions, (b) river sediment with benzoate as a sole carbon source, at a final concentration of 4 mM, incubated under anaerobic conditions (experiment 1) and, (c) BTEX contaminated sediment with toluene as a sole carbon source, at a final concentration of 1 mM, incubated under aerobic conditions, (d) BTEX contaminated sediment with toluene as a sole carbon source, at a final concentration of 1 mM, incubated under anaerobic conditions (experiment 2). In each case incubations were carried out at 30°C .
Chemical analysis
Benzoate degradation was monitored by reverse phase HPLC as previously described [13]. Toluene degradation was monitored by GC/ MS (Agilent) via headspace analysis where samples were injected onto a 105 m × 0.530 mm column (Agilent) and samples were heated to 120°C for 0.5 min followed by a 3°C /min ramp to 140°C and finally a 25°C /min ramp to 250°C holding until completion.
Molecular biology techniques
DNA extraction was carried out, in triplicate, as previously described using bead beating combined with phenol/chloroform extraction [14]. Total microbial DNA was quantified by UV absorption at 260 nm [15]. Primer pair BZAQ4F/R [16] were used to amplify a 484bp fragment from the, α subunit of the bcr gene. The bcr products generated were used to prepare standards for use in qPCR assays with primer BZAQ4F and PFR1 (5`TCCTGMCCGCCSATGTCSAG`3) [17]. Primer PFR1 was designed through multiple sequence alignments (ClustalW-EBI) of bcr α subunits present in T. aromatica, R. palustris, A. evansii and M. magneticum and used to target a small section of the α subunit of the bcr gene for use in qPCR studies. For bdo assays primer pair BDOf and BDOr was used as described previously [18]. All qPCR standards and reactions were performed as described in our previous work [17] using an Opticon 3 real-time PCR thermo cycler (Biorad) coupled with Maxima SYBR Green Master Mix (Fermentas) fluorescent technology.
DNA sequencing was performed by DNA Sequencing & Services (MRCPPU, College of Life Sciences, University of Dundee, Scotland, www.dnaseq.co.uk) using Applied Bio systems Big-Dye Ver 3.1 chemistry on an Applied Bio systems model 3730 automated capillary DNA sequences.
Results
Experiment 1-microbial led degradation of benzoate
In the aerobic, river sediment, microcosm studies complete degradation of benzoate occurred within 3 days of experiment initiation (from an initial concentration of 4 mM). In contrast to this ~20% of benzoate remained in the corresponding anaerobic microcosms following incubation for 14 days (Table 1) upon which point the experiment was terminated. Heat killed controls, maintained ≥ 90% of the initial benzoate added to microcosms under both aerobic and anaerobic conditions. Estimated degradation rates of carbon sources in these microcosms were as follows:
Aerobic benzoate-1.30 ± 0.12 mM.d-1; and
Anaerobic benzoate-0.23 mM ± 0.02.d-1
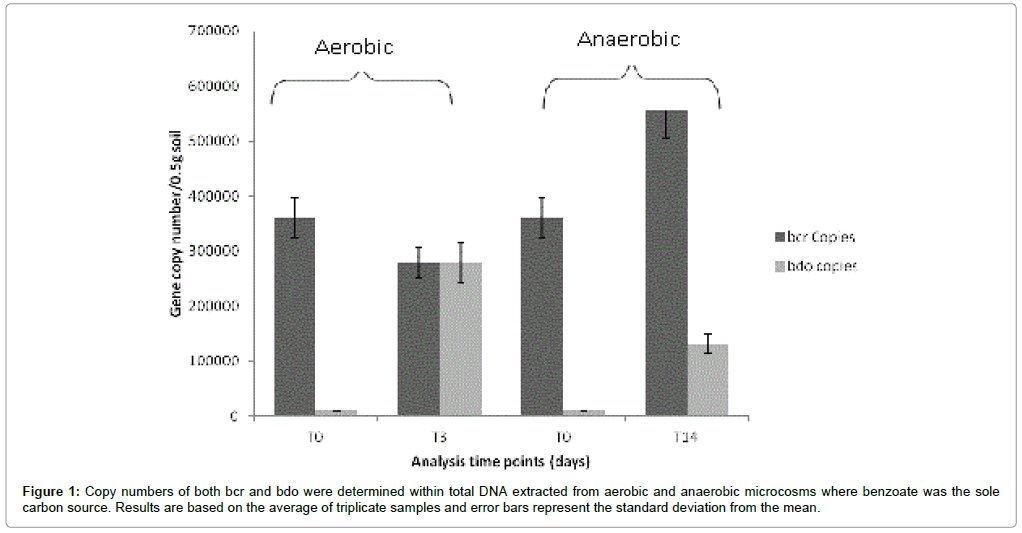
Following incubation both the aerobic and anaerobic microcosms were subjected to DNA extraction followed by further analysis of total DNA. It was observed that qPCR analysis on DNA isolated from benzoate microcosms (Figure 1) indicated that total bcr copies were higher following incubation under anaerobic conditions (5.62×105 ± 3.88×104) in comparison to incubation under aerobic conditions (3.26 × 105 ± 1.9 × 104). In contrast to this observation bdo copies were lower under anaerobic conditions (1.25×105 ± 1.22×104) compared to incubations under aerobic conditions (2.81×105 ± 2.23×104). Thus differences in gene abundances were clearly influenced by the aerobic/ anaerobic conditions of incubation as the same starting inoculum was used for both aerobic and anaerobic microcosms.
Experiment 2-Microbial led degradation of Toluene
In the aerobic, petroleum site sediment, microcosms toluene was completely degraded in under 3 days. In the corresponding anaerobic microcosms ~20% of toluene remained after 6 days (Table 1). In the heat sterilised controls ≥80% of the toluene added to the microcosms remained at the end of the test period under both aerobic and anaerobic conditions. Estimated degradation rates of toluene were determined as follows:-
Aerobic toluene-0.40 ± 0.00 mM.d-1; and,
Anaerobic, toluene-0.13 ± 0.05 mM.d-1
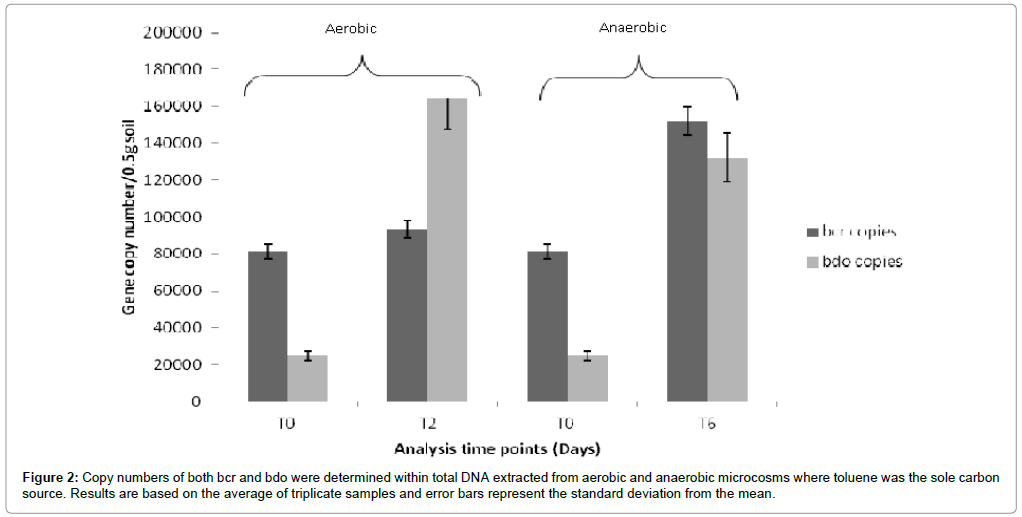
Further qPCR analysis on DNA isolated from toluene microcosms (Figure 2) indicated that bcr copies were higher (1.52×105 ± 6.2×103) following incubation under anaerobic conditions compared to incubation under aerobic conditions (9.3×104 ± 3.2×103). In contrast the bdo copies were lower following incubation under anaerobic conditions (1.32×105 ± 1.65×104) compared to aerobic conditions (1.64×105 ± 1.08×104). In both experiments measurements were made based upon triplicate microcosms. Efficiency of qPCR reactions ranged from101-107% falling within the acceptable range of 90-110% [19].
PCR product sequencing
The nature of the qPCR Products was confirmed through gel electrophoresis and melting curve analysis as described previously [20]. Products were cloned and sequenced to confirm their identity. In each case 20 plasmids were analysed and found to contain inserts of the correct size, 4 of which were randomly picked for sequence analysis. Sequencing indicated that bcr genes obtained from anaerobic benzoate microcosms, using primer pair PFF1/R1, exhibited similarities to Thauera spp (1 sequence), Geobacter spp (2 sequences) and Alkaliphilus spp (1 sequence) bcr genes with homologies ranging from 88-90%.
Similarly bdo genes obtained from aerobic benzoate microcosms shared homologies ranging from 85-87% to Acinetobacter spp (2 sequences) and Pseudomonas spp (2 sequences) bdo genes (accession numbers JQ774497-JQ774500).
Discussion
Microbial degradation rates in microcosms amended with benzoate, under both aerobic and anaerobic conditions, were higher than those rates observed in microcosms amended with toluene. This fact may well be a result of the toxicity of toluene towards microorganisms [21] thus limiting the availability of the compound to a smaller subsection of the overall microbial population present within the toluene microcosms. Unsurprisingly aerobic rates of compound degradation were observed to be higher than anaerobic rates, regardless of the aromatic compound tested.
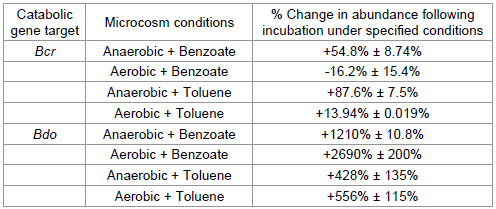
The overall change in the bcr and bdo gene abundances from experimental start up (t zero) to experimental completion (t final) in both the benzoate and toluene microcosms, inoculated with river sediment and sediment from the petroleum site respectively, are documented in Table 2. In both the benzoate and toluene microcosms the catabolic genes tested in this study responded to the redox conditions of each microcosm as the degradation of the target compound proceeded. Under anaerobic conditions, bcr copy numbers increase by a larger percentage over the duration of each experiment relative to aerobic microcosms. In contrast, bdo copy numbers increase by a larger percentage in aerobic microcosms compared to their anaerobic counterparts. This data indicates that comparisons, through qPCR approaches, of the bcr and bdo catabolic genes could be used as a diagnostic tool to differentiate between areas of aerobic and anaerobic bioremediation potential in the subsurface of a polluted site. This data also suggests that studies centred on copy number observations of either bcr or bdo independently would fail to divulge information relating to the aerobic/anaerobic nature of a given site under investigation.
Table 2: Gene abundances upon experimental completion relative to time zero prior to incubation. Each gene studied under various conditions is shown. This data suggests a clear trend in both the benzoate and toluene microcosms, whereby under anaerobic conditions a greater increase in bcr copy numbers is observed, whereas under aerobic conditions a greater increase in bdo copy number is observed.
Results from this work also indicate that more bcr copies were detected overall within the benzoate microcosms compared to the toluene microcosms. This could reflect the differences in the types of microorganisms present in the two inocula. For example microorganisms present in the river sediment used in the benzoate microcosms may experience constantly fluctuating redox conditions as a result of river levels rising and dropping [22] resulting in conditions where facultative microorganisms, which harbour the class I type of bcr, may flourish. In contrast with the conditions which may occur in the river sediment the BTEX contaminated sediment used to construct the toluene microcosms may not have been exposed to oxygen at all for a prolonged period due to its depth and lack of aeration meaning a strictly anaerobic population predominated, capable of degrading toluene through a class II type of bcr enzyme absent from facultative microorganisms [23]. The primers used to detect the bcr gene in this study were designed to target the class I type of bcr enzyme therefore potentially overlooking the strictly anaerobic microorganisms present in both samples. Additional tests using a more definitive range of carbon sources coupled with other sediment types would be helpful in validating this hypothesis.
The microorganisms most closely related to the sequences obtained from this work are well documented in terms of their ability to degrade aromatic hydrocarbons [24] with both Thaura spp Geobacter spp previously being shown to degrade aromatic compounds under anaerobic conditions [25,26].
Conclusion
The results of this work indicate that the copy number of bcr and bdo within microbial communities may be influenced by prevailing redox conditions. This, coupled with copy number changes of each gene, could be useful in determining whether (i) aromatic compound degradation is actively occurring within a given sample; and (ii) degradation of aromatics is occurring under aerobic or anaerobic conditions.
The technique could be used to provide supporting evidence for in situ biodegradation (natural attenuation) and also in the design of in situ remediation systems (e.g. involving oxygen or nitrate/sulphate injection).
Acknowledgements
This work was funded as part of a CAST award from the Department of Education in association with the QUESTOR centre Belfast. Thanks are also extended to Dr Mike Spence for sample donation.
References
- Carmona M, Zamarro MT, Blázquez B, Durante-RodrÃguez G, Juárez JF, et al. (2009) Anaerobic catabolism of aromatic compounds: a genetic and genomic view. Microbiol Mol Biol Rev 73: 71-133.
- Harwood CS, Burchhardt G, Herrmann H, Fuchs G (1999) Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. Fems Microbiol Rev 22: 439-458.
- Chen YD, Barker JF, Gui L (2008) A strategy for aromatic hydrocarbon bioremediation under anaerobic conditions and the impacts of ethanol: a microcosm study. J Contam Hydrol 96: 17-31.
- Fuchs G (2008) anaerobic metabolism of aromatic compounds. Ann N Y Acad Sci 1125: 82-99.
- Gibson DT, Parales RE (2000) Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr Opin Biotechnol 11: 236-243.
- Wilson LP, Bouwer EJ (1997) Biodegradation of aromatic compounds under mixed oxygen/denitrifying conditions: a review. J Ind Microbiol Biotechnol 18: 116-130.
- Flanagan PV, Kelleher BP, Allen CCR (2013) Assessment of anaerobic biodegradation of aromatic hydrocarbons: The impact of molecular biology approaches. Geomicrobiology Journal
- Fuchs G, Boll M, Heider J (2011) Microbial degradation of aromatic compounds - from one strategy to four. Nat Rev Microbiol 9: 803-816.
- Higashioka Y, Kojima H, Sato S, Fukui M (2009) Microbial community analysis at crude oil-contaminated soils targeting the 16S ribosomal RNA, xylM, C23O, and bcr genes. J Appl Microbiol 107: 126-135.
- Powell SM, Ferguson SH, Snape I, Siciliano SD (2006) Fertilization stimulates anaerobic fuel degradation of antarctic soils by denitrifying microorganisms. Environ Sci Technol 40: 2011-2017.
- Beller HR, Kane SR, Legler TC, Alvarez PJ (2002) A real-time polymerase chain reaction method for monitoring anaerobic, hydrocarbon-degrading bacteria based on a catabolic gene. Environ Sci Technol 36: 3977-3984.
- Evans PJ, Mang DT, Young LY (1991) Degradation of toluene and m-xylene and transformation of o-xylene by denitrifying enrichment cultures. Appl Environ Microbiol 57: 450-454.
- Fairley DJ, Boyd DR, Sharma ND, Allen CC, Morgan P, et al. (2002) Aerobic metabolism of 4-hydroxybenzoic acid in Archaea via an unusual pathway involving an intramolecular migration (NIH shift). Appl Environ Microbiol 68: 6246-6255.
- Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ (2000) Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol 66: 5488-5491.
- Sambrook J, Zhou T, Rhankin L (1989) Molecular Cloning: a laborotory manual.
- Song B, Ward BB (2005) Genetic diversity of benzoyl coenzyme A reductase genes detected in denitrifying isolates and estuarine sediment communities. Appl Environ Microbiol 71: 2036-2045.
- Flanagan, Paul V, Kelleher BP, O’Reilly, Szpak SS, et al. (2013) A depth resolved insight into benzoyl CoA reductase and benzoate dioxygenase gene copy numbers within a marine sediment associated with methane seepage. Open Journal of Marine Science: In Press.
- Kim D, Kim SW, Choi KY, Lee JS, Kim E (2008) Molecular cloning and functional characterization of the genes encoding benzoate and p-hydroxybenzoate degradation by the halophilic Chromohalobacter sp. strain HS-2. FEMS Microbiol Lett 280: 235-241.
- Rebrikov DV, Trofimov DY (2005) Real-Time PCR? A Review of Approaches to Data Analysis. Appl Biochem Micro 42: 520-528.
- Kouduka M, Suko T, Morono Y, Inagaki F, Ito K, et al. (2012) A new DNA extraction method by controlled alkaline treatments from consolidated subsurface sediments. FEMS Microbiol Lett 326: 47-54.
- Segura A, Duque E, Mosqueda G, Ramos JL, Junker F (1999) Multiple responses of gram-negative bacteria to organic solvents. Environ Microbiol 1: 191-198.
- Calmano W, Hong J (1993) Binding and mobilization of heavy metals in contaminated sediments affected by pH and redox potential. Wat Sci Tech 28: 223-235.
- Löffler C, Kuntze K, Vazquez JR, Rugor A, Kung JW, et al. (2011) Occurrence, genes and expression of the W/Se-containing class II benzoyl-coenzyme A reductases in anaerobic bacteria. Environ Microbiol 13: 696-709.
- Fritsche W (2006) Aerobic Degradation by microorganisms. Bio/technology, pp. 145-167.
- Boll M, Fuchs G, Heider J (2002) Anaerobic oxidation of aromatic compounds and hydrocarbons. Curr Opin Chem Biol 6: 604-611.
- Wischgoll S, Heintz D, Peters F, Erxleben A, Sarnighausen E, et al. (2005) Gene clusters involved in anaerobic benzoate degradation of Geobacter metallireducens. Mol Microbiol 58: 1238-1252.
Citation: Flanagan PV, Kulakov LA, Kulakova AN, Spence M, Allen CCR (2015) Dynamic Changes in Aromatic Hydrocarbon Associated Catabolic Gene Profiles Linked to Aerobic and Anaerobic Microcosm Studies. Innovative Energy Policies 4: 111. DOI: 10.4172/2090-5009.1000111
Copyright: ©2015 Flanagan PV, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 15505
- [From(publication date): 2-2015 - Dec 19, 2024]
- Breakdown by view type
- HTML page views: 11085
- PDF downloads: 4420