Short Communication Open Access
DNA-protection by Polyamines in Lactobacillus sakei No.2
Jae-ho Choi, Ki-sok Kim, Hyung-ki Do and Cher-won Hwang*AGEE (Advanced Green Energy and Environment), Handong Global University, Pohang, Republic of Korea
- *Corresponding Author:
- Cher-won Hwang
AGEE (Advanced Green Energy and Environment)
Handong Global University
Pohang 791-708, Republic of Korea
Tel: 82 1045332085
E-mail: chowon@handong.edu
Received Date: November 24, 2014; Accepted Date: December 31, 2014; Published Date: January 05, 2015
Citation: Choi J, Kim K, Do H, Hwang C (2015) DNA-protection by Polyamines in Lactobacillus sakei No.2. J Clin Exp Pathol 5:203. doi: 10.4172/2161-0681.1000203
Copyright: ©2015 Choi J, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Clinical & Experimental Pathology
Abstract
In a previous study we showed that we had isolated Lactic Acid Bacteria (LAB) that produce high amounts of the polyamines found in fermented food that function as antioxidants . In this study, we concentrated on another function by investigating polyamines in relation to anti-cancer effects. Although a QR (Quinone Reductase) test showed no anti-cancer effect, good results were obtained for DNA protection and we compared polyamine production and growth of No.2 strain using a RT-PCR machine. This DNA protection may be related to an anti-cancer effect.
After suggesting that polyamines had the ability to protect DNA, we collected polyamines of LAB (Lactobacillus sp. No.2) by HPLC with a fraction collector. Next, Ornithine Decarboxylase (ODC), which is considered as a polyamine producing enzyme, was detected in Lactobacillus sp. No.2 using Western blot and RT-PCR analyses. The results of RT-PCR showed that polyamine production from No.2 strain was related to growth time and especially that culture for 36 hours provided the best conditions for polyamine production. In summary this means that ODC gene expression is related to polyamine production and hence DNA protection.
Keywords
DNA protection; Lactic acid bacteria; ODC; Polyamine
Introduction
Biogenic amines, including tyramine, histamine, putrescine, cadaverine, spermine, and spermidine, can be produced and degraded by the normal metabolic activities of animals, plants, and microorganisms. These amines are mainly produced by microbial decarboxylation of amino acids in foods [1,2].
Putrescine, spermidine, and spermine are three well-known polyamines that are present in the cells of all organisms. Polyamines play an important role in regulating cell growth and proliferation, stabilization of the negative charges of DNA, RNA transcription, protein synthesis, apoptosis, and regulation of the immune response [3]. Polyamines are mostly obtained from dietary sources, and foods containing polyamines can be expected to have cell regeneration and antioxidant effects [3]; however, there are few studies of the polyamine contents of specific strains of microbes that are used as foods.
In this study, we investigated the characteristics of polyamines from the No. 2 strain isolated in a previous study [1] and determined the effectiveness of polyamines (especially spermidine) in DNA protection. Using RT-PCR, it was determined that the production of polyamine from ornithine by Ornithine Decarboxylase (ODC) protein was proportional to the growth of this strain.
Materials and Methods
Polyamine preparation: Polyamines used (putrescine, spermidine, spermine) were standard goods obtained from Sigma-Aldrich and polyamine substances obtained from Lactobacillus sakei sp. No.2 by HPLC (Ultimate 3000, Dionex, USA) [1], with a fraction collector (Foxy Jr., Dionex, USA).
Gel electrophoresis for detecting DNA protection: Polyamines in Lactobacillus sakei sp. No. 2 were collected with a fraction collector from peaks with HPLC. These polyamines and standard polyamines were run through electrophoresis (Mupid-EXu, Japan). DNA protective activity was determined by using a DNA damage assay that was carried out using pBR322. The reaction mixture was prepared by pouring 5 ml of stock solution of the test compound at a concentration of 3000 ppm in an Eppendorf micro test tube. Then 3 ml of diluted plasmid DNA was transferred to the Eppendorf tube. Stock solutions were at 1000 ppm concentrations in the final reaction mixture. Next 3 ml of 2 mM FeSO4 and 4 ml of 30% H2O2 were added successively to each tube. As a positive control, pBR322 DNA with FeSO4 and H2O2 was used. The reaction mixture was incubated for 1 h. After incubation, plasmid DNA was run on 0.9% agarose gel in the dark at 37°C. Agarose (0.9 g) was transferred to a 250 ml flask and then dissolved in 100 ml of 1X TAE (Tris/Acetate/EDTA) buffer and heated in a microwave oven to obtain a homogeneous mixture. After slight cooling to a bearable temperature, 20 ml of the staining agent ethidium bromide was added to the agarose. After gentle shaking, the agarose was poured in a gel tray and allowed to solidify for 30 min. After solidification the 1X TAE (gel running buffer) was poured to such a level that both of the electrodes were submerged in buffer. Then 3 ml of bromophenol blue (loading dye) was added to each reaction mixture after incubation and the samples were loaded on 0.9% agarose gel wells containing TAE buffer. Each reaction mixture with controls was run horizontally in TAE buffer at 100 V and 30 mA for 1 h in an electrophoresis apparatus. The gels were photographed under UV light. For each run, a molecular marker, a negative control, H2O2 treatment, FeSO4 treatment, and test concentrations of CME (Culture Medium Extract) were loaded [4].
RT-PCR for ODC production: Total RNA extraction, first-strand cDNA synthesis, and real-time PCR analysis were performed as described by [5] with some modifications [6]. First-strand cDNA was synthesized with 0.6 μg of total RNA. Primers for target genes were designed using Primer Blast from NCBI and are presented by name and function. Samples were analyzed in a 7500 Real-Time PCR System (Applied Bioscience, Carlsbad, CA, USA) following initial preheating at 95°C for 2 min. Then 40 PCR cycles were performed as follows: 95°C for 15 seconds and 55°C for 30 seconds. The melting curve profiling was conducted at the end of each PCR process to confirm amplification of specific transcripts by the following steps: 95°C for 15 seconds, cooling to 60°C for 1 minute, heating the sample to 95°C for 30 seconds, and cooling to 60°C for 15 seconds under continuous fluorescence measurement [7].
Statistical analysis: The data were analysed by a one-way Analysis of Variance (ANOVA).
Results and Discussion
In this study, we focused on the DNA protection effect of polyamines in Lactobacillus sakei sp. No.2. The polyamines were confirmed by HPLC with a fraction collector from culture medium. Gel electrophoresis was carried out to determine the DNA-protection properties of the polyamines, and they were compared to standard polyamines.
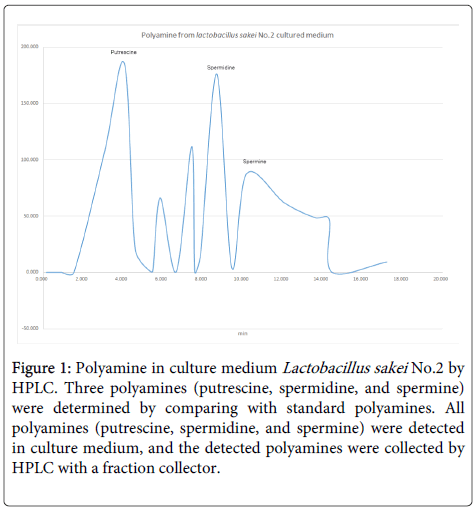
Analysis of polyamines in No.2 strain: We determined and collected polyamines in No.2 strain [1] culture medium to compare with standard polyamines (Sigma-Aldrich, USA). Large amounts of putrescine and spermidine were detected from the culture medium by HPLC (determined by standard polyamines) and were collected by a fraction collector (Figure 1).
Figure 1: Polyamine in culture medium Lactobacillus sakei No.2 by HPLC. Three polyamines (putrescine, spermidine, and spermine) were determined by comparing with standard polyamines. All polyamines (putrescine, spermidine, and spermine) were detected in culture medium, and the detected polyamines were collected by HPLC with a fraction collector.
DNA protection analysis by gel-electrophoresis: To consider one of the many functions of polyamines, the original purpose of this study was to define the anti-cancer effect of polyamines from No.2 strain as described in previous papers [8,9]. A QR (Quinone Reductase) test failed to detect any anti-cancer activity of the polyamines from No.2 strain (data not shown); therefore, we focused on their cancer prevention ability through DNA protection.
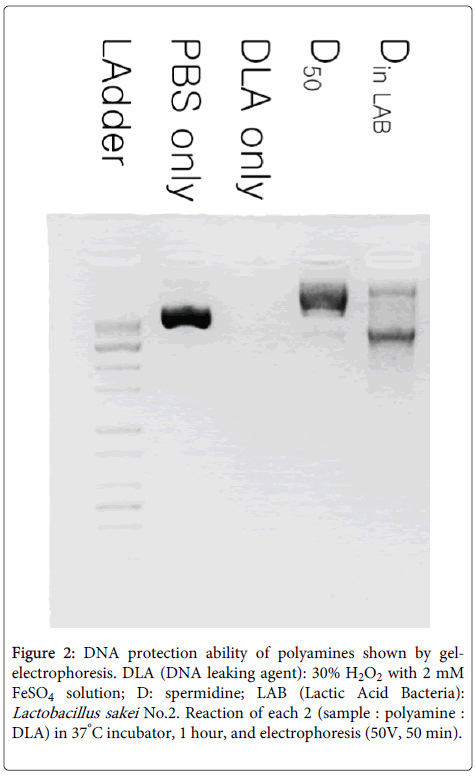
Medium cultured for 36 hours was prepared because this provides the best conditions for producing polyamines in No.2 strain. The relationship between DNA protection and polyamines from No.2 strain was investigated and no detectable levels of DNA protection could be found for putrescine and spermine, but there was a relationship between spermidine and DNA protection (Figure 2).
Figure 2: DNA protection ability of polyamines shown by gelelectrophoresis. DLA (DNA leaking agent): 30% H2O2 with 2 mM FeSO4 solution; D: spermidine; LAB (Lactic Acid Bacteria): Lactobacillus sakei No.2. Reaction of each 2 (sample : polyamine : DLA) in 37°C incubator, 1 hour, and electrophoresis (50V, 50 min).
We think that polyamines have a pre-anticancer effect because they can protect DNA and prevent cancer cells from developing.
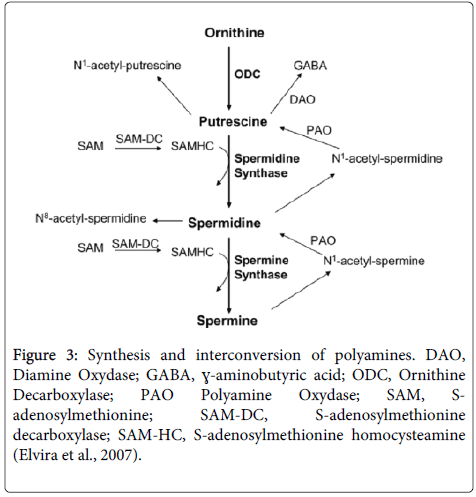
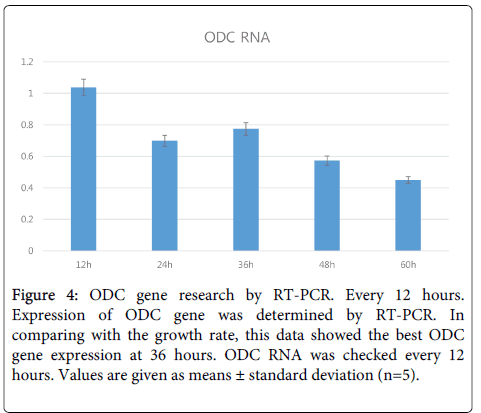
ODC gene expression analysis and mRNA level by real-time PCR: After suggesting that sperimidine is related with DNA protection, we investigated the Ornithine Decarboxylase (ODC) production ability of the No.2 strain. Polyamines from ornithine catalyzed by ODC are produced in the order of putrescine, spermidine, and spermine [3], and ODC protein is the starting invertase for producing polyamines as shown in Figure 3. We searched for the ODC sequence from Lactic Acid Bacteria (LAB) with the odcf/odcr (5’- TGCACTTCCATATCCTCCAG/5’-GAATTTCTGGAGCAAATCCA, 127-bp amplicon) primers [10]. This data showed that ODC is best revealed at 36 hours, which is consistent with the results of previous work that determined the best time in culture medium for polyamine content (Figure 4). These results for ODC content are very similar to the growth rates of No.2 strain.
Conclusion
This study suggests that polyamines from No.2 strain have a DNA protection function and prove that the ODC gene in this strain is related with polyamine production. Polyamines are known for DNA protection. Polyamines interact with DNA phosphate groups by means of nonspecific electrostatic bonds [11]. This interaction has been shown to result in the protection of small DNA molecules from common damaging agents, such as ionizing radiation and reactive oxygen species. Polyamines in solution with polynucleotides have also been shown to inhibit the activity of endonucleases, including DNase I [11].
We determined with a HPLC fraction collector the polyamines in No.2 strain culture medium and with a qRT-PCR system that they were produced by ODC. Even though sperimine showed limited DNA protection ability in a gel electrophoresis experiment (data not shown), the results showed that spermidine from No.2 strain is a good candidate for use as a functional material. Through our study, we believe that spermidine from No.2 strain has the potential to be developed as a functional material for DNA protection with a high commercial value; however, further studies are required, including additional tests on the spermidine synthase gene in No.2 strain.
Acknowledgment
This research was supported by the Handong Global University Research Grants No. 20130088.
References
- Choi J, Hwang C, Do HK, Kwon GS, Kang SC (2013) Anti-oxidizing Functional Effect of Polyamine as a Bioconversion Starter Using Microbes Isolated from Fluke babsikhae. J Korean SocApplBiolChem 56: 83-86.
- Brink B, Damink C, Joosten J, Huisin’t Veld J (1990) Occurrence and formation of biologically active amines in food. Int J Food Microbiol 11: 73–84.
- Larqué E, Sabater-Molina M, Zamora S (2007) Biological significance of dietary polyamines. Nutrition 23: 87–95.
- Abbas S, Hussain M, Ali S, Parvez M, Raza A, et al. (2013) Structural, enzyme inhibition, antibacterial and DNA protection studies of organotin (IV) derivatives of thiophene-2 -carboxylic acid. Journal of Organometallic Chemistry 724: 255-261.
- Choi BH, Ahn IS, Kim YH, Park JW, Lee SY, et al. (2006) Berberine reduces the expression of adipogenic enzymes and inflammatory molecules of 3T3-L1 adipocyte. ExpMol Med 38: 599-605.
- Ji Y, Kim HN, Park HJ, Lee JE, Yeo SY, et al. (2012) Modulation of the murine microbiome with a concomitant anti-obesity effect by Lactobacillus rhamnosus GG and Lactobacillus sakei NR28. Beneficial. Microbes 3: 13-22
- Kim K, Choi J, YosepJi, Park S, Do H, et al. (2014) Impact of bubble size on growth and CO2 uptake of Arthrospira (Spirulina) platensis KMMCC CY-007. Bioresource Technology 170: 310–315
- Huang Y, Hager ER, Phillips DL, Dunn VR, Hacker A, et al. (2003) A Novel Polyamine Analog Inhibits Growth and Induces Apoptosis in Human Breast Cancer Cells. Clinical Cancer Research 9: 2769–2777.
- Wallace HM, Fraser AV (2003) Polyamine analogues as anticancer drugs. Biochemical Society Transactions 31.
- Nannelli F, Claisse O, Gindreau E, de Revel G, Lonvaud-Funel A et al. (2008) Determination of lactic acid bacteria producing biogenicamines in wine by quantitative PCR methods. Letters in Applied Microbiology.
- Agostino LD, Pietro M, Luccia AD (2005) Nuclear aggregates of polyamines are supramolecular structures that play a crucial role in genomic DNA protection and conformation. FEBS Journal 272: 3777–3787.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 14316
- [From(publication date):
February-2015 - Apr 02, 2025] - Breakdown by view type
- HTML page views : 9755
- PDF downloads : 4561