Mini Review Open Access
DNA-Lipids-Me2+ Complexes Structure and their Possible Functions in a Cell
Kuvichkin VV*
Institute of Cell Biophysics, Russian academy of Sciences, Russia
- *Corresponding Author:
- Kuvichkin VV
Institute of Cell Biophysics
Russian academy of Sciences
142290, Pushchino, Moscow reg. Russia
Tel: +7-916-640-5267
Fax: +7-4967330509
E-mail: vvkuvichkin@gmail.com
Received date: November 23, 2015; Accepted date: January 5, 2016;Published date: January 12, 2016
Citation: Kuvichkin VV (2016) DNA-Lipids-Me2+ Complexes Structure and their Possible Functions in a Cell. J Chem Biol Ther 1:105. doi: 10.4172/2572-0406.1000105
Copyright: © 2016 Kuvichkin VV. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Chemical Biology & Therapeutics
Abstract
Study of lipid-nucleic acids interactions have fifty years of history and were originally aimed at studying of DNAmembrane complexes (DMC). On the basis of own and literary data we had been proposed DMC model and their participation in the formation of structures nucleoid and nuclear envelope with pores.
Understanding of nuclear pores role not only as channels between nucleus and cytoplasm but also as basic regulator of gene expression will allow using this knowledge in nanobiotechnology and medicine, in particular in gene therapy. In gene therapy the basic method of genes transfer in cells are complexes of DNA with which are strongly toxic for man. Zwitterionic lipids compose to 50 weight % of membranes and besides participation in DMC formation can really substitute cationic surfactants as carrier of DNA in cells.
Introduction
Three types of interactions exist between main cell molecules, proteins, nucleic acids and lipids: protein - nucleic acids, protein- lipid and lipid - nucleic acids. Lipid - nucleic acids interactions are wide studied in the world on last 30 years. Successful using of lipids and liposome in gene therapy for gene delivery to cells and organs promoted development of these researches. As the cellular membranes more than on 50% consist of lipids, the interaction DNA with membranes in many respects is caused by lipid - nucleic acids interactions.
An idea about the possible involvement of bacterial membranes in cellular genome functioning has appeared for the first time in 1963. The first data on DNA binding to a membrane were obtained by electron microscopy. In 1963 Jacob et al. suggested the involvement of DNA-membrane complexes in chromosome segregation between daughter cells [1]. In this work, the role of lipids in the attachment of DNA to a membrane not mentioned. However, data on electron microscopy that provided evidence on numerous DNA–membrane complexes (further DMC) in vivo [2], have forced some scientists toward research, focused on the study of the DNA–lipid interactions in vivo and in vitro .
The subsequent burst of biochemical investigation of DNAmembrane complexes (DMC) in 1965 -1985 was reviewed in [3,4]. These investigations gave many important discoveries, which included the evidence of DNA involvement in the initiation of transcription and replication, in the formation of the structure of bacterial and eukaryotic nucleoid.
Based on our own and literature data we proposed a DMC model and the involvement of DMC in the formation of the structure of nucleoid, Bayer’s junctions, nuclear matrix, nuclear envelope with pores.
The understanding of a role of nuclear pores not only as channels between nuclei and cytoplasm, but also as a main regulator of gene activity makes us possible to use this knowledge in nanotechnology and medicine, in particular in gene therapy.
In gene therapy the main technique for a transfer of genes into the cell are DNA complexes with cationic surfactants which are highly toxic for humans.
Zwitterionic phospholipids account for up to 50 Wt % of membranes and along with the involvement of DMC formation they may actually compete with cationic surfactants for a role of DNA carrier. A variety of DNA- zwitterionic lipids- Mg2+ complexes depending on their component concentration is shown. The use of DNA-lipid-Mg nanoparticles of the known structure enables to construct nonharmful for the humans, non-virus vectors to treat different diseases.
DNA-lipid complexes in vitro
Manzoli et al. [5,6] reported considerable changes of parameters of DNA melting as a consequence of interaction of DNA with various lipids dissolved in 70% ethanol solution. The ethanol was used for better solubility of lipids.
Lipids are only weakly soluble in water, but form micelles or liposomes. These particles, however, cause intensive light scattering, which represent major limit for application of light spectroscopy methods. Therefore, the results of the study of DNA–liposome complexes by spectroscopy methods were ambiguous. Manzoli et al. showed both stabilizing and denaturing effect of various lipids on DNA. Experiments with DNA–liposomes complexes revealed a depletion of DNA melting peak when DNA molecules were inside the liposomes [7]. IR- spectroscopic analysis of DNA–lipid complexes (total fraction of lipids of a rat liver) showed existence of DNA denaturation at the presence of these lipids [8].
The data of calorimetric study (insensitive to changes of light scattering) of the complexes: polyA*polyU-phosphatidylcholine liposomes in absence of divalent cations have shown minor changes of profile in the melting of lipid and polynucleotide [9]. Thus, the interactions of DNA with liposomes from zwitterionic or negatively charged phospholipids do not cause to a significant changes of the structure components of complexes, except some special methods of preparation of complexes (using organic solvents [10], dryingrehydration, and placement of DNA inside liposomes [7]. Only interaction of positively charged natural lipid sphingosin with DNA results in significant changes of structure of lipid bilayer [11], but this lipid present in a cell in very low concentrations.
Lipids of chromatin
Lipids are present in many cellular structures containing DNA and proteins, for instance, in nuclear matrix [12], nucleoid [13] and chromatin [14-20]. Literature provides more data on the presence of lipids in chromatin as compared to those in nuclear matrix and nucleoid. But still, it is a matter of critical discussion regarding artifact origin of lipids in chromatin composition.
The so-called lipids tightly bound to chromatin have been explored for more than forty years [21]. The authors suggested to isolate chromatin DNA of maximum molecular weight, i.e., practically intact chromosome. After phenol extraction at which almost all proteins were removed, it was found out that the lipids in amount of 10% remained tightly bound to chromatin, herewith, the composition of lipids depended on physiological state of the cell. A regulatory role of lipids was assumed in transcription and replication processes.
Despite our positive attitude to this hypothesis in our first model of DMC [22], DNA directly interacts with lipids in the area of nuclear pore annulus we have to discuss in a way of critical thinking the origin of lipids tightly bound to chromatin. The use of organic solvent, phenol, in the course of isolation of the mentioned lipids is disputable.
It is known that lipids interact with chromatin in the area of nuclear pores of eukaryotes or “Bayer’s junction” prokaryotes [23]. Probably these lipids differ in composition from the total lipids of nuclear or bacterial membrane. During isolation of nucleoid these lipids transfer to nucleoid core, but after lysozyme- detergent treatment some lipids could be removed from nucleoid, so the lipids that remain in the nucleoid composition are distinguished by the composition from the lipid composition in DMC. We can also hypothesize that in the presence of phenol some lipids are removed from nuclear pore and composition of lipids tightly bound to chromatin could significantly differ from the lipid composition in nuclear pores. The method that enables to estimate the lipid composition in nuclear pore is planned to develop taking the method of isolation of “membrane DNA”, recently suggested [24] as a basis.
The results of study performed at the laboratory headed by Albi (1983-2009) could undergo similar critics whereas the detergent is used during isolation of lipids tightly bound to chromatin. However, the composition of these lipids differ from the composition of chromatin lipids obtained at the laboratory of Strazhevskaya. Using biophysical methods of localization of lipid domains in nucleus Albi and Magni showed significant difference of these lipids from total fractions of nuclear lipids. The relation of these lipids to transcription and replication processes could be interpreted by their close associations with DMC (nuclear pores and transcription factories). It has been demonstrated that in hepatocyte nuclei the chromatin phospholipid fraction is localized near the RNA in decondensed chromatin [18]. The necessity of developing new method of isolation of lipids tightly bound to chromatin (DMC) without using organic solvent and detergent is still relevant for these authors.
Almost simultaneous with the start of the research of DNA–lipid interactions in vitro , Budker et al. [25] have revealed the existence of specific interactions between zwitterionic phospholipids and polynucleotides at the presence of divalent metal cations (Me2+).
Role of multivalent cations in the interaction of DNA with phospholipids
During the study of DNA-phosphatidylcholine liposomes-Mg2+ complexes Budker et al. [25] have detected, that DNA can interact with liposomes composed of zwitterionic liposomes by means of a bridge from divalent cations. An extensive study of triple or ternary complexes (TC) has been performed by BudkerVG, Kuvichkin VV, Bergelson LD, Zhdanov RI, Suleymanoglu E, Balgavy P et al. in 1978-2015.
The brief summary of the results of the study of TC is listed below:
• DNA forms TC with three main lipids: phosphatidylcholine (PC), phosphatidylethanolamine (PE) and sphingomyeline (SM), the addition of other lipids increases or decreases these interactions [26].
• Ability of divalent cations to form TC correlates with a degree of binding of these cations with PC [26].
• The liposomes fuse partially or completely at presence of Me2+, and DNA [26].
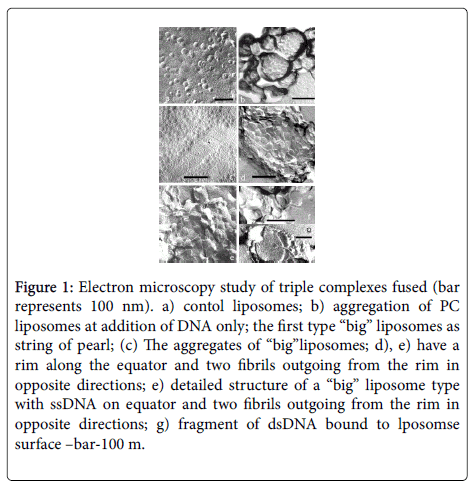
• DNA acts as a fusogen, untwisting in the zone of liposome fusion. We have shown the fact of partial fusion of liposomes, along with untwisted DNA regions, by technique of freeze-etching and cryo- TEM (Figure 1), [27,28].
Figure 1: Electron microscopy study of triple complexes fused (bar represents 100 nm). a) contol liposomes; b) aggregation of PC liposomes at addition of DNA only; the first type “big” liposomes as string of pearl; (c) The aggregates of “big”liposomes; d), e) have a rim along the equator and two fibrils outgoing from the rim in opposite directions; e) detailed structure of a “big” liposome type with ssDNA on equator and two fibrils outgoing from the rim in opposite directions; g) fragment of dsDNA bound to lposomse surface –bar-100 m.
• DNA molecules in TC become restrictedly accessible to action of DNAse I, but are more accessible to digestion of S1-endonuclease [29].
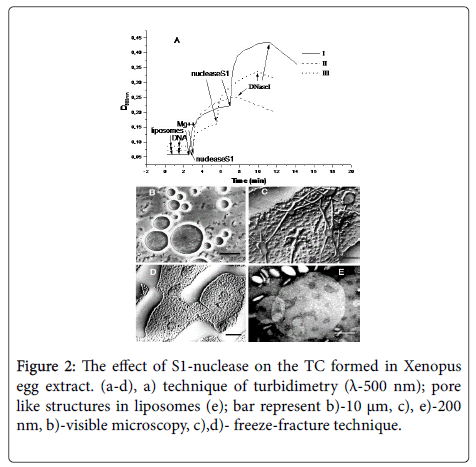
• The treatment of TC formed in Xenopus eggs extract with S1- nuclease– an enzyme that digests of ssDNA – results in a complete fusion of liposomes with the formation of “giant” liposomes up to 10 μm in diameter [29], (Figure 2b).
• The method of turbidimetry does not distinguish fusion from aggregation, but nevertheless is able to give an idea of how S1- nuclease affects triple complexes. So, after the complexes have been formed, the addition of S1-nuclease initiates further increase in the optical density of a sample [29].
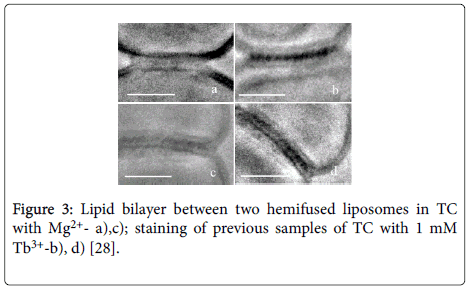
• Using Cryo-TEM method ssDNA presence was confirmed in the area of fusion of two DOPC liposomes induced by Mg2+ (Figures 3a and 3c) with subsequent staining of samples a),c) by Tb3+ (Figures 3b and 3d).
Figure 3: Lipid bilayer between two hemifused liposomes in TC with Mg2+- a),c); staining of previous samples of TC with 1 mM Tb3+-b), d) [28].
• The staining of TC with Tb3+ which preferentially binds to ssDNA supports our TC model where DNA double helix unwinds in TC.
• By means of 31P-NMR technique a strong change of signal connected with phosphate groups of PC was showed, that evidence about the formation of complexes between phospholipids, DNA and Mg2+ [30,31].
• A cooperative character of binding of Mn2+ in the complex DNA - PC - was shown [32];
• Three-stranded polynucleotide polyA *2 poly U forms TC too as revealed by micro- calorimetry [9].
Binding energy between phospholipid and DNA molecules by means of bridges of Mg2+ is comparable with that of hydrogen bounds between the bases of DNA (~ 2-3 kcal/mole per one base pair).
DNA-membrane complexes: early studies
As we mentioned above, the first data on DNA binding to a membrane were obtained by electron microscopy [2]. Results of study of DMC were reviewed in a number of works [3,4,33].
Bayer [23] has shown that in bacteria there exist so-called zones of adhesion between a bacterial wall and cytoplasm membrane, to which DNA is attached. There exist from 100 to 400 of such contacts in a cell. It has been assumed, that the site of DNA attachment to a nuclear membrane of eucaryotes are the pores [27,33]. In 1970-80 the attempt was made to isolate a fraction of DMC and to analyze its biochemical composition. For this purpose the cells have been gently lysed in presence of Mg-sarcosylate and so-called "M-band" fraction has been obtained [34]. This fraction contained DNA, enriched by moderate repeats, newly synthesized DNA, RNA and proteins [35,36]. There is no information on lipid composition of "M-band" fraction. The involvement of DMC in an initiation of replication and transcription and in a regulation of gene expression has been suggested [37]. Further research as well as improvement of the methods of isolation of DMC fractions has not been continued mostly due to a general fascination by study of nuclear matrix [12].
In connection with a present topic, it is necessary to consider a problem of structure of bacterial and eucaryotic nucleoid, those are directly related both to DMC and to the nuclear matrix.
DMC and nucleoid of bacteria and eucaryo
The problem of packaging of giant DNA (1-2 cm in length) in bacteria, whose size does not exceed a several microns, is comparable with complexity of packaging DNA in eucaryotic chromosomes. After a mild lysozyme - detergent lysis of bacteria the structure called as a nucleoid (“chromosome”) of bacteria was detected by electron microscope [13]. As soon as the nucleoid have been prepared from eucaryotic nucleus [37] the assumption was made, that inside the bacteria there exist same compact structure, as in chromosomes of eucaryotes. However electron microscopy of bacteria did not reveal any structures similar to chromosomes like structures and no central core nucleoid containing outer and inner membrane of bacteria as in a "membrane bound" nucleoid . We developed model of formation of bacterial nucleoid with participation of DNA-membrane complexes, and model of the DNA-membrane contacts [38,39].
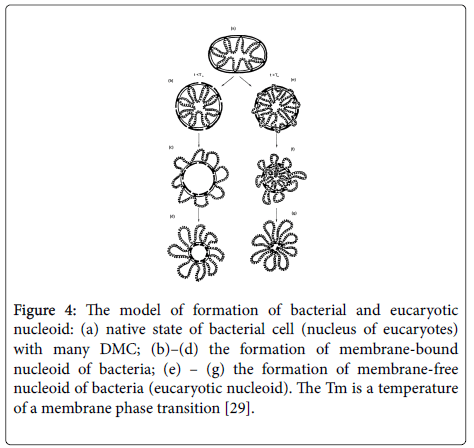
The essence of this model is in the following. Both in a native bacterial cell, and in an interphase nucleus of eucaryotes, DNA is attached to a membrane in many points. This arrangement is the basis of further structure of nucleoid. This structure can arise from treatment of a nucleus or bacterial cell by detergents and removing major part of the membrane with preserving some part of DMC. As a result of this process a small membrane island with closed loops of attached DNA appears. This structure, i.e., nucleoid, is observed in electron microscope; however it does not exist in a native cell (Figure 4).
Figure 4: The model of formation of bacterial and eucaryotic nucleoid: (a) native state of bacterial cell (nucleus of eucaryotes) with many DMC; (b)–(d) the formation of membrane-bound nucleoid of bacteria; (e) – (g) the formation of membrane-free nucleoid of bacteria (eucaryotic nucleoid). The Tm is a temperature of a membrane phase transition [29].
According to our opinion, the similarity of a nucleoid of bacteria and eucaryotes consists in fact that DNA in an interphase nucleus and dividing cells of bacteria is attached to a membrane in a number of sites (Figure 3a). This condition is necessary for normal transcription and replication. In addition, the structure of DMC and the method of its formation in bacteria and eucaryotes are according to our model similar. According to this model DNA is attached to a membrane at socalled regulatory sites (promotors, sites of initiation of replication, which according to reassociation kinetics belongs to the moderate repeats). We suggest that at DNA-membrane binding sites are also localized low molecular RNA (l mw RNA).
These RNA form a triple helix with DNA, which due to its interaction with a membrane lipids unwind already at a room temperature and forms a classical R-loop: DNA-RNA hybrid and a single-stranded DNA [39]. In contrast to a double helix DNA the three-stranded hybrids unwind at lower temperature, and thus could be easily attached to a membrane. We showed, that three-stranded nucleic acids can form complexes with PC-liposomes, which results in their unwinding already at a room temperature [9]. It is known, that a low molecular weight RNA (l mw RNA) is able to bound with a chromatin and could form three-stranded helix. The role of l mw RNA in a regulation of transcription was earlier reported [40].
The data on relaxation of the nucleoid’s structure induced by RNAseH and S1-nuclease- enzymes, that destroys the DNA-RNA hybrids and ssDNA respectively, confirm the opinion on participation of triple helix structures on DNA attachment to a membrane [41].
There is an opinion that nuclear pores and "Bayer's junction" are the sites of attachment of DNA to a membrane. The DNA sites between pores or "Bayer's junction", are, according to our model, replicons. However, number of loops in a nucleoid is less than number of replicons owing to particular destruction of DMC by detergents during the isolation of nucleoids.
Nuclear pore model
Nuclear pore for the current data is a complex of more than thirty proteins- nucleoporins. The first models of the nuclear pore were based on immuno-electron microscopy methods of the position nucleoporins determination in the pore complex is not very accurate. The task was complicated by the fact that the composition nucleoporins pore complex in various organisms has been different but transport functions such pores were similar. Moreover, the composition nucleoporins in the nuclei of cells of various tissues was also varied. This made it possible to add nucleoporins function of controls for genome expression, but greatly complicates the creation of a uniform model of the nuclear pore on the basis of some proteins.
In accordance with our new nuclear pore model [41] DMC arise in the chromatin areas with three-stranded hybrids: DNA - low molecular weight RNA (l mw RNA) at their interactions with two membrane vesicles (~70 nm in diameter). The melting temperature of DNA/l mw RNA triple helix is considerably lower than the same sequence of double-stranded DNA. That can specify preferential attachment of triple-stranded hybrids to the membrane vesicles.
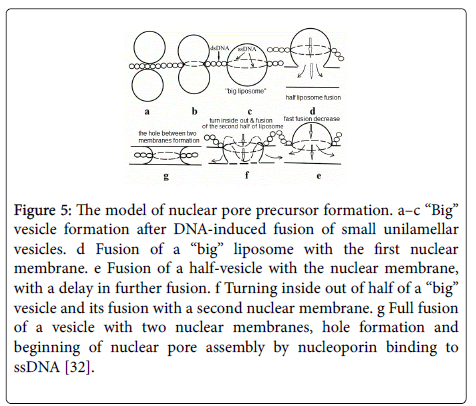
The triple helix unwinding during fusion of two nuclear membrane vesicles results in pre-pore structure: DNA/l mw RNA hybrid and a single-stranded DNA (ssDNA) located on the outer diameter of fused “big vesicle” (Figure 5c).
Figure 5: The model of nuclear pore precursor formation. a–c ‘‘Big’’ vesicle formation after DNA-induced fusion of small unilamellar vesicles. d Fusion of a ‘‘big’’ liposome with the first nuclear membrane. e Fusion of a half-vesicle with the nuclear membrane, with a delay in further fusion. f Turning inside out of half of a ‘‘big’’ vesicle and its fusion with a second nuclear membrane. g Full fusion of a vesicle with two nuclear membranes, hole formation and beginning of nuclear pore assembly by nucleoporin binding to ssDNA [32].
The “big vesicle” during interaction with double nuclear membrane can form nuclear pore channel, fusing two membranes. During this fusion ssDNA and hybrid DNA/l mw RNA shifts to annulus of pore.
Nuclear membranes originate from endoplasmic reticulum or vesicles (200 nm in diameter) not forming TC with DNA and Me2+. Nuclear pore assembly has several stages and involves surface tension of nuclear membranes and “big” vesicle. The nucleoporins interacting with ssDNA and dsDNA /l mw RNA hybrid in pore annules form the final structure of the pore complex.
‘‘Anaphase’’ and ‘‘Interphase’’ nuclear pore
Some ‘‘big’’ vesicles, bound earlier to chromatin, are unable to fuse with an inner nuclear membrane because of weak decondensation of this chromatin fraction. As a result of DNA replication in S phase,decondensation of the same chromatin areas increases and vesicles bound to this chromatin fraction will be able to reach the nuclear membrane and create additional ‘‘interphase’’ pores. This could explain the second peak of nuclear pore formation in S phase [42]. These pores may differ from ‘‘anaphase’’ pores in nucleoporin composition and in the dynamics of pore complex assembly. The reason could be depletion of nucleoporins stock in the nucleus or the use of nucleoporins in the construction of intranuclear annulate lamellae.
Nuclear pores and regulation of genome expression
Pre-pores are the result of two equal membrane vesicles fusion induced by DNA located between the vesicles containing mainly zwitterionic lipids in the presence of Mg2+ [4]. Then two membrane vesicles fuse to form one ‘‘big’’ vesicle, belted at the equator by a ssDNA and DNA/RNA hybrid. As we thought previously, these low molecular weight RNAs are snRNA U1- and U2-type, (161 and 193 nucleotides in size), respectively [22,41]
The term ‘‘big’’ for a vesicle should be used very conditionally, since the diameter fused liposome is only 1.26 times larger than initial liposome diameter. During DNA-induced vesicle fusion, the DNA/l mw RNA triplex unwind into an ssDNA and double hybrid DNA/l mw RNA because triplex thermal stability is much lower than that of pure DNA. Besides, l mw RNA determines the specificity of certain regions of DNA attachment to the nuclear envelope and regions of ssDNA are the ideal sites for transcription initiation. We try to give an answer to the question: “If DNA is bound to the pore complex, then why DNA is invisible in the area of nuclear pores in modern EM images of cellular nuclei?” Naked DNA is hardly visible even on the surface of liposome with the use of cryo-TEM high resolution method, but it is actually impossible to view it on the surface of nuclear membrane which contains proteins and lipids.
TC and transcription factories
The bulk of the nuclear pores are formed in anaphase, about the same quantity in interphase. What happens to the TC, which had not reached the nuclear envelope and remains in the nucleoplasm. Not all pre-pores reach the nuclear envelope and form nuclear pores during chromosome decondensation in prophase. Pre-pores remaining inside of the nucleus form aggregates, which may be the transcription factories discovered not so long ago in the nuclei [43]. These structures are very similar to pre-pore aggregates and are transcriptional active. They are very similar to the TC structure studied by us earlier [4]. If you look at a ssDNA in the nuclear pore complex annule and ssDNA encircling the big liposome in TC, they are located in similar lipid membrane environment, except the curvature of the surface, which is at the TC and in pore complex annule has the opposite sign. This implies that ssDNA of TC and ssDNA in a pore annule are good transcription initiation sites. In the works Hozak [44] shows intranuclear structure with high transcriptional activity and structural units resembling TC. We hope that more detailed analysis of transcriptional factories by methods high resolution microscopy should show the validity of our hypothesis. Other centers of transcriptional activity are linear clusters of nuclear pores [24].
A pleasant surprise for the author was publication of new article of Dr. E Suleymanoglu which supplements our knowledge about properties of TC and their possible use in pharmacy [45].
Nuclear pores and pharmacy
Nuclear pores with a variety of function should be under the pharmacy spotlight. They could be the target for different substances with low and high molecular weight. As minimum, a test for binding of any medical preparation to nuclear pores is necessary before it can be used in medical practice. The simplest way of performing this test is a check of binding ability of drugs with TC.
References
- Jacob F, Brenner S,Cuzin F (1963) On the regulation of DNA replication in bacteria. Cold Spring HarborSymp Quant Biol 28: 329-347.
- Ganesan A, Lederberg J (1965) A cell-membrane bound fraction of DNA. BiochemBiophys Res Commun18: 824-835.
- Moyer MP (1979) The association of DNA and RNA with membranes.Int Rev Cytol 6: 1-61.
- Kuvichkin VV (2002) Lipid-nucleic acids interactions in vitro and in vivo.Bioelectrochemistry 58: 3-12
- Manzoli F, Muchmore J, Bonora B, Sabioni A, Stefoni S (1972) Interaction between sphingomyelin and DNA. BiochimBiophysActa 277: 251-255.
- Manzoli F, Muchmore J, Bonora B, Capitani S, Bartoli S(1974) Lipid-DNAinteractions. II. Phospholipids, cholesterol, glycerophosphorylcholine, spingosine and fatty acids. BiochimBiophysActa 340: 1-15.
- Brosius B, Riesner D (1986) Influence of lipid membranes on the conformational transitions of nucleic acids. J BiomolStructDyn4: 271-290.
- Shabarchina L, Sukhorukov B, Kuvichkin V (1979) Infrared spectroscopic study of DNA-lipid interactions. DNA compacting on dispersed particles.Biofizika 24: 990-998.
- KuvichkinV, Emeljanenko V, KuznetsovaS, ZhdanovR, Petrov A (1999) Calorimetric study of the complexes: polyA*polyU-phosphatidylcholineliposomes-Mg2+. Biophysics 44: 386-394.
- Budker V, Slattum P, Monahan S, Wolff J (2002) Entrapment and Condensation of DNA in Neutral Reverse Micelles. Biophys J 82: 1570–1579.
- Kõiv A, Mustonen P, Kinnunen P (1994) Differential scanning calorimetry study on the binding of nucleic acids to dimyristoylphosphatidylcholine -sphingosine liposomes. Chemistry and Physics of Lipids 70: 1–10.
- Berezney R (1991) The nuclear matrix: A heuristic model for investigating genomic organization and function in the cell nucleus.JCell Biochem47:109-123.
- Pettijohn DE (1982) Structure and properties of the bacterial nucleoid. Cell 30: 667-669.
- Struchkov VA, Strazevskaya NB (2000) Structural and functional aspects of nuclear lipids of normal and tumor cells. Biochimiya 65: 620-643.
- Struchkov VA,Strazhevskaya NB, Zhdanov RI (2002) Specific natural DNA-bound lipids in post-genome era. The lipid conception of chromatin organization. Bioelectrochemistry 56: 195-198.
- Viola Magni MP, Gahan PB,Albi E, Iapoce R, Gentilucci PF (1985) Chromatin phospholipids and DNA synthesis in hepatic cells. Basic ApplHistochem 29: 253-259.
- Viola Magni MP, Gahan PB, Pacy J (1985) Phospholipid in plant and animal chromatin. Cell Biochem. Funct 3: 71-78.
- Albi E, Micheli M, Viola Magni MP (1996) Phospholipids and nuclear RNA. Cell BiolInt 20: 407-412.
- Albi E, Viola Magni MP (2004) The role of intranuclear lipids. Biol Cell 96: 657-667.
- Albi E,Lazzarini A,Lazzarini R,Floridi A, Damaskopoulou E, et al. (2013) Nuclear Lipid Microdomain as Place of Interaction between Sphingomyelin and DNA during Liver Regeneration.Int J MolSci 14: 6529-6541.
- Beliaev SD, Strazhevskaia NB, Kolomiitseva IK, Struchkov VA, Kuzin AM(1974)DoklAkadNauk SSSR Lipids in a high molecular weight complex of rat thymus and liver DNA. Russian 214:1189-1191.
- Kuvichkin V (1983) Theoretical model of DNA-membrane contacts.Biofizika 28: 771-775.
- Bayer ME (1979) Bacterial outer membranes: Biogenesis and functions. Wiley and Sons, N.Y.
- Kuvichkin VV (2012) Isolation of chromatin DNA tightly bound to the nuclear envelope ofHeLa cells. J MembrBiol 245: 683-690.
- Budker V , Godovikov A, Naumova L, Slepneva I (1980) Interaction of polynucleotides with natural and model membranes. Nucleic Acids Res 8: 2499-2515.
- Kuvichkin V, Sukhomudrenko A (1987) Interaction of natural and synthetic polynucleotides with liposomes in the presence of divalent cations. Biofizika 32: 628–633.
- Kuvichkin VV. (1990) Ultrastructural study of DNA--liposomes--Mg2+ complexes. Biofizika 35: 256-262.
- Kuvichkin V, Danev R, Shigematsu H, Nagayama K (2009) DNA-induced aggregation and fusion of phosphatidylcholine liposomes in the presence of multivalent cations observed by the cryo-TEM technique. J MembrBiol 227: 95-103.
- Kuvichkin V, Uteshev V (2003) The role of lipid-nucleic acids interactions in nuclear envelope and pore complex assembly. Biophysics 48: 236-239.
- Viktorov A, Grepachevskii A, Bergel'son L (1984) DNA-phospholipid interaction. 31P-NMR study. BioorgKhim 10: 935-939.
- Zhdanov R, Volkova L, Rodin V (1994) Lipid Spin Labeling and NMRStudy of Interaction Between Polyadenylic Acid: Polyuridylic Acid Duplex and Egg PhosphatidylcholineLiposomes. Evidence for Involvement of Surface Groups of Bilayer. Phosphoryl Groups and Metal.ApplMagnReson7: 131-146.
- Kuvichkin V, Volkova L, Naryschkina E, Isangalin F (1989) ESR-studyof complexes DNA- PC-liposomes-Mg2+Biophysics 34: 405- 409.
- Kuvichkin VV (1982) The role of membranes in organization and expression of eukaryote genome. Dep.in VINITI 09.04.82, ? 2199-82. Moscow, p68.
- Comings D, Okada T (1973) DNA replication and the nuclear membrane.J MolBiol 75: 609-618.
- Harmon J, Taber H (1977) Some properties of a membrane-deoxyribonucleic acid complex isolated from Bacillus subtilis. J Bacteriol 129: 789-795.
- Hobart P,Duncan R, Infante A (1977) Association of DNA synthesis with the nuclear membrane in sea urchin embryos. Nature 267: 542-544.
- Winston S, and Sueoka N (1980) DNA-membrane association is necessary for initiation of chromosomal and plasmid replication in Bacillus subtilis (initiation mutants/chromosomal origin/plasmid). ProcNatlAcadSci USA 77: 2834-2838.
- Benyajati C, Worcel A (1976) Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster.Cell 9: 393–407.
- Sukhorukov B, Kuvichkin V, Shabarchina L (1980) Structure and function of DNA--membrane contact in cells. Biofizika 25: 270-275.
- Petukhova O, Mittenberg A, Kulichkova V, Kozhukharova I, ErmolaevaIu, et al. (1997) A new class of small RNP (alpha-RNP) containing antisense RNA in K-562 cells. IV. The coordinated regulation of the expression of Alu-containing mRNA and alpha-RNA during differentiation. Ontogenez 28: 437-444.
- Kuvichkin V (2011) The mechanism of a nuclear pore assembly: a molecular biophysics view.J MembrBiol 241: 109-116.
- D'Angelo M,Hetzer M (2008) Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol18: 456-466.
- Yildirim S, Castano E, Sobol M, PhilimonenkoV, Dzijak R, et al. (2013) Involvement of phosphatidylinositol 4,5-bisphosphate in RNA polymerase I transcription. J Cell Sci126: 2730–2739.
- Sobol M, Yildirim S, Philimonenko V, Marášek P, Castaño E, et al. (2013)UBF complexes with phosphatidylinositol 4,5-bisphosphate in nucleolar organizer regions regardless of ongoing RNA polymerase I activity.Nucleus4: 478-486.
- Funda F,Demirsoy K, Eruygur N,Suleymanoglu E (2015) Supramolecular Langmuir monolayers and multilayered vesicles of self-assembling DNA–lipid surface structures and their further implications in polyelectrolyte-based celltransfections. J Nanopart Res17: 50.
Relevant Topics
- Analytical Methods
- Biocatalysis and biotransformation
- Bioinorganic Chemistry
- Biological chemistry
- Bioorganic chemistry
- Chemical Biology of Tetracyclines
- Chemotherapeutic Agents
- Enzyme Inhibitor
- Mechanisms of DNA Damage and Repair
- Nanomaterials For Imaging and Drug Delivery
- Natural Product Biosynthesis
- Nucleic Acid Analogs
- Spectroscopic Probes
Recommended Journals
Article Tools
Article Usage
- Total views: 13971
- [From(publication date):
February-2016 - Apr 18, 2025] - Breakdown by view type
- HTML page views : 13029
- PDF downloads : 942