DNA Methylation and Mutator Genes in Escherichia Coli K-12
Received: 06-Jun-2022 / Manuscript No. bcp-22-67437 / Editor assigned: 09-Jun-2022 / PreQC No. bcp-22-67437 / Reviewed: 14-Jun-2022 / QC No. bcp-22-67437 / Revised: 20-Jun-2022 / Manuscript No. bcp-22-67437 / Published Date: 27-Jun-2022 DOI: 10.4172/2168-9652.1000382
Abstract
Mutator strains of Escherichia coli have been used to define mechanisms that account for the high fidelity of chromosome duplication and chromosome stability. Mutant strains defective in post-replicative mismatch repair display a strong mutator phenotype consistent with a role for correction of mismatches arising from replication errors. Inactivation of the gene (dam) encoding DNA adenine methyltransferase results in a mutator phenotype consistent with a role for DNA methylation in strand discrimination during mismatch repair. This review gives a personal perspective on the discovery of dam mutants in E. coli and their relationship to mismatch repair and mutator phenotypes
Keywords
Alkylating agents; DNA repair; Escherichia coli; Mutator; Methylation; Recombination
Introduction
Chromosome replication requires a high degree of fidelity, and studies in Escherichia coli K-12 over the last fifty years or so have identified the major mechanisms by which this is achieved. The experimental approach used to solve the fidelity question has relied mainly on the isolation and characterization of mutator strains. A mutator phenotype (Mut) is displayed by mutants that have an increased spontaneous mutation frequency relative to wild type (Mut+). The underlying assumption is that such bacteria are impaired in systems that normally correct spontaneous replication errors and, in general, this assumption has been correct. It took some time, however, for this assumption to take hold given that the first E. coli mutator strain was described in 1954 [1] and systematic screening for mutator strains did not begin until 1970 [2].
In this review I have focused on a group of related mutator strains (and one in particular) that has been the subject of my research for the past few decades. I decided to present a personal view of the developments in this research area in the hope it offers insight into the history of these mutants and will be more entertaining than a formal scientific summary. The latter part of the review is a more conventional summary of mutator genes and their effects, and further details can be found in other reviews [3,4]. Genes discussed in this review are listed with a brief explanation of each (Table 1).
| Gene | Gene product |
|---|---|
| dam | DNA adenine methyltransferase |
| dcm | DNA cytosine methyltransferase |
| dnaB | Replicative helicase |
| dnaE | Catalytic alpha-subunit of DNA polymerase III holoenzyme |
| dnaG | DNA primase |
| dnaQ | Epsilon-subunit of DNA polymerase III holoenzyme |
| fpg | Synonym for MutM |
| hexAB | MutSL homologs |
| lig | DNA ligase |
| mutD | Allele of dnaQ resulting in defective proofreading |
| mutH | Mismatch repair endonuclease |
| mutL | Mismatch repair protein |
| mutM | Glycosylase specific for oxidized guanine-cytosine basepairs |
| mutS | Detects base mispairs to initiate mismatch repair |
| mutT | Prevents incorporation of oxidized guanine into DNA |
| mutU | Allele of uvrD |
| mutY | Glycosylase specific for oxidized guanine-adenine basepairs |
| polA | DNA polymerase I |
| polC | Synonym for dnaE |
| recA | Promotes synapsis of homologous DNA strands |
| recBCD | Double-strand end-specific exonuclease |
| ruvA | With RuvB acts as a Holliday junction translocase |
| ruvB | With RuvA acts as a Holliday junction translocase |
| ruvC | Holliday junction resolvase |
| uvrD | Mismatch repair associated helicase |
Table 1: E. coli genes considered in this article.
DNA methylation mutants
I was appointed to my first faculty position as an Instructor at Rutgers Medical School (as it was known then) in Piscataway, NJ, in 1971. I had come to join N. Ronald Morris who had been researching DNA methylation in eukaryotes. It seemed worthwhile studying the problem in E. coli which, unlike eukaryotes, had both 6-methyladenine (6-meA) and 5-methylcytosine (5-meC) in its DNA. The approach would be a standard one – isolate mutants lacking methylated bases and deduce their functions by examining their properties.
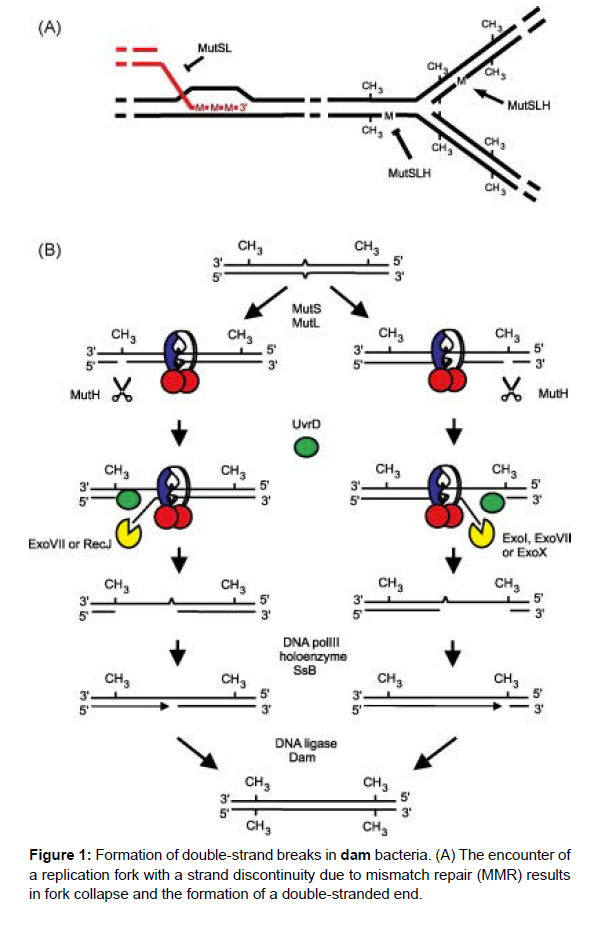
DNA sequence analysis has shown that 5-meC is involved in generating single base changes spontaneously This appears to occur through spontaneous deamina tion of 5-meC to yield thymine. The G/T mismatch upon DNA replication would result in a G-C to A-T base pair change. Deamination of cytosine is not mutagenic, since a specific repair system excises uracil, the deamination product. Evidence for a role for 5-meC residues in genetic recombination has been obtained. One piece of evidence is that dcm mutations suppress the hyper-rec phenotype of arlbacteria. The nature of the interaction between arl and dcm genes and/or products is not clear at present but are under intense investigation. Another role for 5-meC residues might be to alter protein- DNA interactions. That is, a protein may bind differentially to a specific region of DNA depending on the state of DNA methylation. Evidence for this has recently been obtained. The lexA repressor binds less well to DNA if the binding site does not contain its single 5-meC residue [5]. All dam mutations, except one, map in a single complementation group and are recessive to the wild type allele. In contrast to dcm mutants, dam strains show a variety of phenotypic traits. These include increased spontaneous mutability; increased sensitivity to certain alkylating agents, base analogs and ultra-violet (UV) light; a hyper-rec phenotype; increased induction of lysogenic bacteriophages; inviability of dam- ~-, dam- recB-, dam- recC- and damlexA- double mutants and suppression of some dam phenotypic traits by mutL-, mutS-, sinand uvrn”. These phenotypes a~ot present in dam+ revertants of dam mutants. The pleiotropy of phenotypes suggests that 6-meA and/or the dam methylase have several biological roles in cellular metabolism. A model which accounts for some of the phenotypes of dam- strains has been proposed. It supposes that 6-meA residues determine strand specificity for repair of mismatched bases. That is, the unmethylated strand of a duplex DNA containing a mismatch is subject to excision, whereas the other methylated strand is not. This model can account for the mutability phenotype; the sensitivity to base analogs and the indirect suppression of dam- by mut- mutations. The mutH, and genes have been shown to be involved in mismatch repair (Figure 1).
Acknowledgement
None
Conflict of Interest
None
References
- Liu F, Zhang X, Zhao B, Tan X, Wang L, Liu X (2021). Role of Food Phytochemicals in the Modulation of Circadian Clocks. J Agric Food Chem 8735-8739.
- Ko SH, Kim HS.(2012) Menopause-Associated Lipid Metabolic Disorders and Foods Beneficial for Postmenopausal Women. Nutri1-110
- Ma L, Sun Z, Zeng Y, Luo M, Yang J (2021). Molecular Mechanism and Health Role of Functional Ingredients in Blueberry for Chronic Disease in Human Beings. Int J Mol Sci 27-85
- Kolb H, Kempf K, Martin S(2020) Health Effects of Coffee: Mechanism Unraveled . Nutri : 18-42.
- Gupta PS, Bhargava SP, Sharma ML (1962) Acute copper sulphatepoisoning with special reference to its management with corticosteroid therapy. J Assoc Physicians India 10: 287-292.
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Citation: Willson J (2022) DNA Methylation and Mutator Genes in Escherichia Coli K-12. Biochem Physiol 11: 382. DOI: 10.4172/2168-9652.1000382
Copyright: © 2022 Willson J. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 1555
- [From(publication date): 0-2022 - Feb 21, 2025]
- Breakdown by view type
- HTML page views: 1336
- PDF downloads: 219