Research Article Open Access
DNA Fingerprinting Based Decoding of Indica Rice (Oryza sativa L) Via Molecular Marker (SSR, ISSR, & RAPD) in Aerobic Condition
| Tissue culture Lab, College of Biotechnology, University of Agriculture & Technology, Meerut, India | ||
| Corresponding Author : | Ashu Singh Tissue culture Lab College of Biotechnology Sardar Vallabh Bhai Patel University of Agriculture and Technology Meerut-250110, India Tel: 9412472292 E-mail: ashubiot25@gmail.com |
|
| Received February 11, 2015; Accepted April 22, 2015; Published April 24, 2015 | ||
| Citation: Singh A, Sengar RS (2015) DNA Fingerprinting Based Decoding of Indica Rice (Oryza sativa L) Via Molecular Marker (SSR, ISSR, & RAPD) in Aerobic Condition. Adv Crop Sci Tech 3:167. doi: 10.4172/2329-8863.1000167 | ||
| Copyright: © 2015 Singh A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | ||
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
||
Visit for more related articles at Advances in Crop Science and Technology
Abstract
Genetic improvement mainly depends on the extent of genetic variability present in the population. The molecular marker is a useful tool for assessing genetic variations and resolving cultivar identities. The objective of this study was to evaluate the genetic divergence of 30 rice varieties (Basmati, Non-Basmati, Aerobic) using 10 ISSR, RAPD markers each. The diversity or similarities and dissimilarities between all thirty rice varieties were calculated using 0 1 sheets. SSR primers RM-263 is highly informative since it recorded high PIC value (0.995). The resolving power varies between 0.132(RM-256) to 4.662(RM-222) with an average value of 2.7502. In RAPD analysis PIC values varies from 0.811(OPD-08) to 0.9925(OPF-13) with average of 0.9635 and resolving power varies from 1.32(OPJ-08) to 2.066(OPJ-13) with average of 1.8256. In ISSR analysis, PIC value ranged from 0.8791(ISSR6) to 0.9916(ISSR5)
with an average value of 0.9482. The resolving power varies between 1.6(ISSR3) and 8.366(ISSR2) with an average value of 5.2708. The PIC values and Resolving power were calculated for individual primers. The analysis indicated that ISSR expressed maximum resolving power of 8.336 and RAPD gave maximum PIC values of 0.9925. RAPD primer OPF-13 gave the maximum accessions coverage (depending on the value of PIC) in the rice genome. Out of 52 amplified bands, 49 bands were polymorphic and 3 bands were monomorphic. The cluster analysis using the marker systems could distinguish the different genotypes. The dendogram generated on the principle of Unweight Pair Wise Method using Arithemetic Average (UPGMA) was constructed by Jaccard’s Coefficient and the genotypes were grouped in to clusters. The dendogram developed for aroma and quality traits showed that the genotypes with common phylogeny and geographical orientation tend to cluster together thus marker based molecular fingerprinting could serve as a sound basis in the identification of genetically distant accessions as well as in the duplicate sorting of the morphologically close accessions as the case is common in differentiating Basmati and non-basmati.
| Keywords | |
| Phylogen; Polymorphism; Indica rice; Luster analysis; cTAB method; Genome coverage; Genetic diversity | |
| Introduction | |
| Rice (Oryza sativa L.) is one of the leading cereal crops of the world and is the principal food crop of about half of the world’s population. It is a major source of calories for them [1]. In many regions, it is eaten with every meal and provides more calories than any other single food. It can also be used in the manufacture of cosmetics and textiles; beer and wine are also made from it [2]. Besides its food value, it has high cultural and social values in rice consuming societies. Rice is the staple food of more than 50% of the world’s population [3]. By the year 2025, 21% increase in rice production will be needed over that of year 2000 [4]. It is one of the most important crops that provide food for more than half of the world population [5]. | |
| This implies that thousands of valuable allelic variations of traits of economic significance remain unutilized [6]. The first step towards determining the magnitude of these risks is to evaluate the genetic diversity in improved rice genotypes as the success of a crop improvement program depends on the magnitude of genetic variability and the extent to which the desirable characters are heritable [7]. Hence assessment of genetic diversity becomes important in establishing relationships among different cultivars [8,9]. Therefore, different rice varieties of distinct genetic structure are a good promise for the future rice crop improvement. Thus, identification of genotypes and their inter-relationships is important. Development of new biotechnological techniques provides increased support to evaluate genetic variation in both phenotypic and genotypic levels and the results derived from analyses of genetic diversity at the DNA level could be used for designing effective breeding programs aiming to broaden the genetic basis of commercially grown varieties. | |
| DNA fingerprinting/profiling is used to describe the combined use of several single locus detection systems and is being used as versatile tools for investigating various aspects of plant genomes including characterization of genetic variability, genome fingerprinting, genome mapping, gene localization, analysis of genome evolution, population genetics, taxonomy, plant breeding [10]. Genetic diversity can be evaluated with morphological traits, seed proteins, isozymes and DNA markers. Molecular marker technology is the powerful tool for determining genetic variation in rice varieties. In contrast to morphological traits, molecular markers can reveal abundant difference among genotypes at the DNA level, providing a more direct, reliable and efficient tool for germplasm characterization, conservation, and management and untouched by environmental influence. In the present study SSR, ISSR and RAPD markers were used, as these are dominant marker systems and are less costly and easier to be developed and used. The genetic diversity analysis can be extended to characters like salt tolerance/ other abiotic stress, which are controlled by large number of QTLs which may share homology between genes responsible for other abiotic stresses like temperature, drought, flood, submergence etc. Refinement in primer sequences and increasing their specificity in relationship to characters under study can enhance the efficacy of microsatellite markers as a tool for tagging specific gene throughout the concerned genome [11]. In the present study, 30 rice varieties were analyzed for genetic variation using SSR, ISSR & RAPD markers. Specially, the objective of the study was DNA fingerprinting and genetic diversity analysis of different varieties (Basmati, nonbasmati & aerobic) to measure the extent of genotypic differences, genetic relationship and to assist in broadening the germplasm base of future aromatic rice breeding programs. | |
| Materials and Methods | |
| The field trial involving thirty Basmati and non-Basmati rice varieties adapted to traditional irrigated and aerobic agro-eco systems of rice cultivation was conducted at Sardar Vallabhbhai Patel University of Agriculture and Technology, Meerut during Kharif (Rainy) crop season in 2012-2013. For molecular studies, genomic DNA was isolated from single leaf taken from each of the 30 varieties/genotype of the rice following CTAB (Cetyl Trim ethyl Ammonium Bromide) method [12]. The 30 varieties/genotypes were subjected to classify for genetic diversity of rice with the help of 10 SSR, 10 ISSR, 10 RAPD primers. Agarose gel electrophoresis was used to quantify DNA on the basis of molecular weight. The purified DNA was amplified in PCR with different SSR, ISSR, RAPD primers (10 each). The 30 varieties/ genotypes were subjected to screen for diversity of Basmati and non- Basmati genotypes adapted to irrigated and aerobic conditions. | |
| For evaluating marker efficiency, PIC (Polymorphism Information Content) value and Resolving powers were estimated for each primer. | |
| Data analysis and detection of genetic diversity for SSR, ISSR & RAPD markers | |
| Thirty one rice varieties were used to estimate genetic diversity. Polymorphic products from all the marker system were assayed for presence (1) or absence (0). The proportion of bands that have been shared between any of the two varieties averaged over loci SSRs ISSRs & RAPDs were used as the measure of similarity. Genetic diversity was calculated using formula given by Chakravarthi et al. [12]. It refers to the value of a marker for detecting poly- morphism within a population, depending on the number of detectable alleles and the distribution of their frequency. | |
| The power of each primer to distinguish among the studied genotypes was evaluated by the resolving power (Rp) [12]. Resolving power is the capacity of any primer to distinguish among different varieties. It is defined per primer as Rp=Σ Ib where lb is the band informativeness, that takes the values of: 1-(2x [0.5-p]), being p the proportion of the rice varieties containing the band. | |
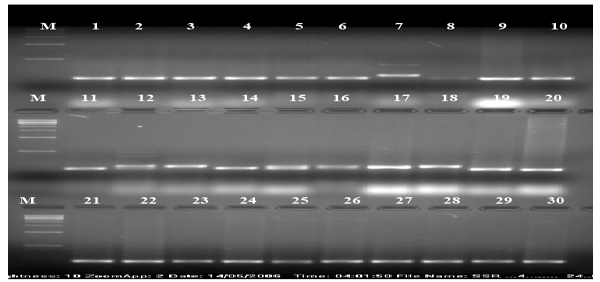
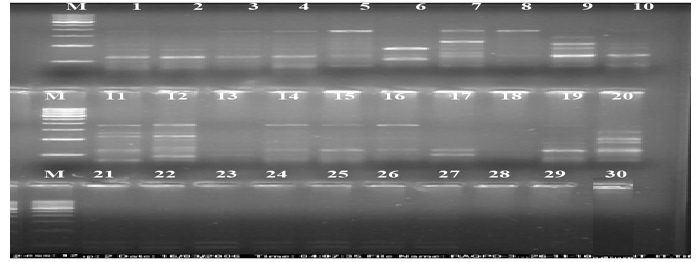
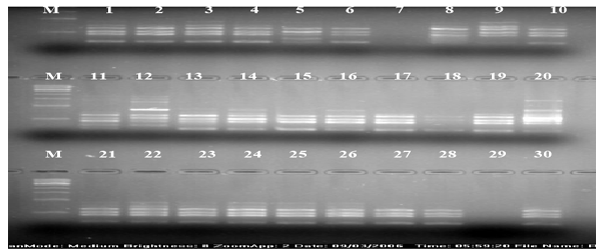
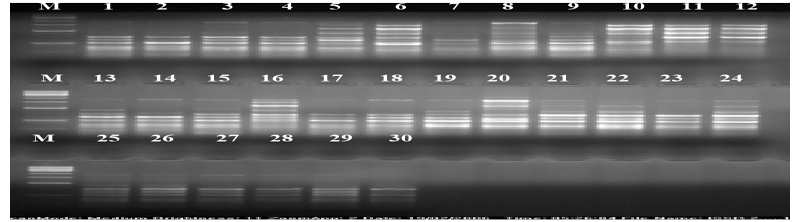
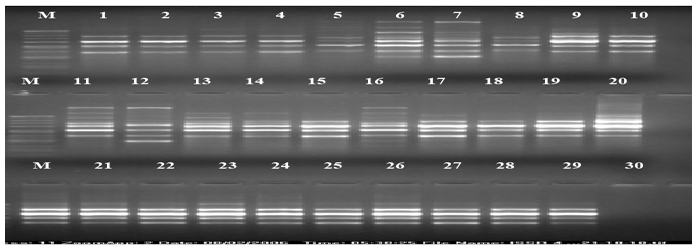
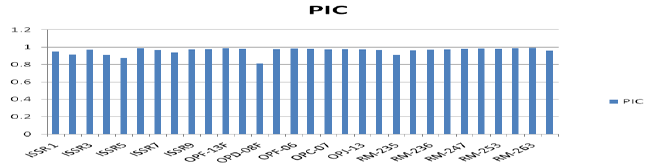
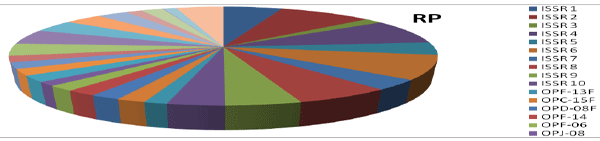
| The calculation was based on the number of bands in SSRs, ISSRs, & RAPD primers. Clustering pattern was based on distance matrices by using the Un- weighted Pair Group Method Analysis (UPGMA) program in NTSYS-pc version 2.2 [13]. Thirty rice varieties were analyzed for Genetic diversity and molecular characterization. Numbers of polymorphic as well as monomorphic bands were obtained for determination of PIC value and resolving power of 30 markers (Table-1). Gel photographs are given in Figures 1-3 for SSR, RAPD & ISSR respectively. The Resolving power of RAPD, ISSR & SSR molecular markers are presented by Pie diagrams (Figure 4) and PIC of combined markers values are given as bar diagram (Figure 5). Comparative analysis of RAPD, ISSR & SSR is given in Table 1. Clustering of genotypes based on different marker assays and joint assays is given in Figure 6 onwards. | |
| Discussion | |
| Traditionally used morphological and chemical parameters have not been found to be discriminative enough, warranting more precise techniques. Presently several molecular techniques are available for fingerprinting different cultivars of crops involving differences within and among cultivars. Among these, the DNA Markers RAPD, simple sequence repeat (SSR) and ISSR markers are considered effective and cost-efficient which could detect higher degree of polymorphism in rice [14]. | |
| In this present study, the detailed use of molecular markers for the assessment of genetic diversity and identification of economically important traits were evaluated. Genome analysis based, molecular markers have generated epitome of information and a number of databases. The availability of new techniques and new equipment such as invention of PCR technology has revolutionized progress of research in molecular biology. PCR-based methods during the last more than 20 years became the routine work of molecular laboratories. Presently publications related to the methodology and applications of PCRbased DNA fingerprinting behave like the DNA in a PCR, i.e., they multiply exponentially. DNA based molecular markers are the most powerful diagnostic tools to detect DNA polymorphisms both at the level of specific loci and at the genome level [15]. | |
| DNA based molecular markers are the most powerful novel tools to detect variation in rice genotypes on the basis of DNA polymorphisms both at the level of specific loci and at the whole genome level [16]. Polymorphisms at the DNA level can be studied by numerous approaches like polymorphism information content etc [17]. Direct strategy is the determination of the nucleotide sequence of a defined region [18], the establishment of lineage of this sequence to an orthologous region in the genome of related organisms. The extent of homology between various sequences can be deduced from the alignment, and phylogenies reconstructed by a variety of approaches and algorithms. DNA sequencing provides highly robust, reproducible, and informative data sets that can be utilized to different analyses for discrimination or mapping of targeted regions of a genome [19]. On the other hand, DNA sequencing can be tedious and expensive when very large number of individuals has to be assayed (e.g., in population genetics and marker-assisted plant breeding programs). In specific areas of research, it is not suitable for estimation of genetic diversity. PCR-based molecular markers SSR (Simple sequence repeat) and ISSR (Inter simple sequence repeat) instead provide a measure of genome wide genetic variation [20]. The analysis of genetic diversity and relatedness between or within different populations, species, and individuals is very important for many disciplines of biological science. Marker technology based on polymorphisms in DNA has catalyzed research in a variety of disciplines such as phylogeny, taxonomy, ecology, genetics, and plant and animal breeding [21]. | |
| Despite such large number of varieties developed using diverse germplasm, molecular marker based diversity analysis has shown the genetic base of Indian rice gene pool to be surprisingly narrow [22,23]. Moreover, with regard to trends of genetic diversity in major Indian rice cultivars, however, little work has been done, recently hypervariable microsatellite markers evenly distributed in rice genome have been demonstrated to be quite effective in estimating genetic diversity [24] and during last three decades, a classical strategy for estimation of genetic variability has been complemented by molecular techniques. These include, for example, the analysis of chemical constituents, but most importantly relate to the development of molecular markers. | |
| SSR analysis | |
| The term SSR (simple sequence repeat) also known as microsatellites was coined by Litt & Luty. These are Co-dominant markers [31]. They are a class of repetitive DNA sequences usually 2.6 bp that are distributed throughout whole genome and are flanked by highly conserved region [32]. The objective of this present study was to evaluate these markers essentially belonging to the repetitive DNA family. Simple sequence repeats (SSRs) consist of 1 to 6 bp long monomer sequences which are repeated several times. A microsatellite fingerprint database, has been generated with 10 SSR markers for 30 rice genotypes, and used for diversity analysis and determination of genetic relationships, as we know the high level of polymorphism associated with microsatellites represents just one component of their rapid rise to become the “genetic tool of choice” for mappers working with all animal as well as plant species. | |
| In SSR analysis, a total of 24 amplified bands were detected using 10 SSR primers in 30 rice varieties, out of 24 amplified bands, only 1 was monomorphic and 23 were polymorphic (Table 2). The maximum numbers of polymorphic bands (4 bands) were obtained using RM- 235 primer with 98% polymorphism. RM-235 observed minimum polymorphism with PIC Value of 0.912, Lower PIC value may be the result of closely related genotypes and higher PIC values might be the result of diverse genotypes. Low PIC values for some other primers were earlier reported by Ma et al. [33]. Among the primers used in the present study, RM-263 is highly informative since it recorded high PIC value (0.995) value of. The resolving power varies between 0.132(RM- 256) to 4.662(RM-222) with an average value of 2.7502. On the basis of PIC values & RP (resolving power) cleared that SSR primers have important position in this analysis. In the analysis of SSR assay, all the 30 rice varieties were classified in four main clusters (Figure 3). All the ten Basmati varieties were clustered together. In addition, MAUB 13 and PS 2511 both the non-Basmati genotypes were grouped in the same cluster. The reason, being that these varieties had common parentage. MAUB-13 a non-Basmati and a Basmati variety Vallabh Basmati-21(Similarity 100%) had the same parentage. All varieties/ genotypes adapted in aerobic conditions in Srilanka were also included in the same cluster along with Sarbati a variety of Indian origin. Two genotypes shahpasan (Chhattisgarh origin) and Sathi, a farmers’ variety (Traditionally conserved in Northern India), both suitable for aerobic condition clustered together in a main cluster. The rest all non-Basmati rice genotypes were clustered together. DG154 and DG138 expressed maximum similarity (approximately 100%) perhaps they had same line of origin or they have been selected on the basis of phenotypic differences those were governed by small DNA sequences with a difference of very few base pairs. Also, DG234 and DG296 had maximum similarity (approximately 100%). | |
| RAPD analysis | |
| Williams et al. used simplest version of molecular markers, the Random Amplified Polymorphic DNA (RAPD). These are mainly Dominant markers [31]. Such primers are simple arbitrary sequences of decamer nucleotides with a GC content of at least 50%. Such primers are used under relaxed stringent conditions and in such cases no prior knowledge of DNA sequence is required. Such primers are still being used despite their low reproducibility. This is due to the simplicity of this technique, as an only very small amount of DNA is required and information on template DNA sequence is not needed [34]. | |
| This study indicates that RAPD is a sensitive and powerful technique to distinguish among rice cultivars and to detect genetic variation at DNA level. The results can complement classical morphological identification. The RAPD technique requires a small amount of DNA extracted from a single rice seed, and is not affected by environmental factors. In contrast, morphological identification involves the evaluation of many morphological parameters of the whole plant, and can be affected by environmental factors. DNA-based analysis could be used to identify Indica rice cultivars to prevent fraudulent commercial activity. However, the RAPD is not suitable for the analysis of DNA extracted from processed products, because of the high degradation rate of DNA in products. | |
| RAPD analysis revealed a large number of distinct, scorable fragments per primer pair. A total of 52 bands were amplified using 10 RAPD primers in 30 rice gentotypes. Out of 52 amplified bands, 49 bands were polymorphic and 3 bands were monomorphic (Table 2). The number of amplified fragments varied from 3 to 49, with an average of 5.2 (i.e. Average no of bands per primer). The PIC values and resolving power were calculated for individual primers. Polymorphic Information Content (PIC) refers to the value of a marker for detecting polymorphism within a population, depending on the number of detectable alleles and the distribution of their frequency whereas resolving power is the capacity of any primer to distinguish among different varieties. | |
| In RAPD analysis, PIC values varies from 0.811(OPD-08) to 0.9925(OPF-13) with average of 0.9635 and resolving power varies from 1.32(OPJ-08) to 2.066(OPJ-13) with average of 1.8256. RAPD markers could be employed both for estimating the relationships between varieties and for variety identification [35,36]. All the 30 rice varieties of two quality groups (Basmati, Non-Basmati) adapted in different agroeco systems were grouped into three distinct major clusters (Figure 3). The Basmati type varieties/genotypes were not clustered together and distributed over three sub clusters in different major clusters. MAUB- 13, MAUB-15 and MAUB-64 were clustered with Taroari Basmati, a Basmati variety suitable for normal as well saline/alkaline conditions. Pusa Basmati-1 was clustered with Basmati-370. Another variety of quality rice PS-1121 was clustered with PS-2511. While MAUB-57 the elite Basmati genotype could not be clustered with any variety. Also, NDR118 and Sathi varieties could not be grouped with other varieties. Therefore, these varieties were put individually in two separate clusters. The major cluster was subdivided into 10 sub clusters in case of RAPD. The genotypes having far distant place of origin were clustered together reflecting that the distribution of varieties over clusters was independent of their place of origin/development. It indicated the uniqueness of the DNA sequences represented by RAPD markers in the whole genome. Maximum varieties of Srilanka origin were grouped in one major cluster; perhaps they had same line of origin or were selected on the basis of minor differences governed by few numbers of base pairs. | |
| ISSR analysis | |
| ISSR (Inter-simple sequence repeat) primers are an alternative form of SSR and can be utilized to amplify inter-SSR DNA sequences [28]. These are mostly dominant markers though occasionally a few of them exhibit co-dominance. A total of 71 bands were amplified using 10 ISSR primers in 30 rice varieties. Out of 71 amplified bands, 7 bands were monomorphic and 64 were polymorphic (Table 2). In ISSR analysis, PIC value ranged from 0.8791(ISSR6) to 0.9916(ISSR5) with an average value of 0.9482. The resolving power varies between 1.6(ISSR3) and 8.366(ISSR2) with an average value of 5.2708. | |
| In ISSR analysis, all 30 rice varieties classified in two main clusters (Figure 3). Sathi separately clustered with the Srilanka rice varieties. The export quality variety Pusa1121 was clustered with varieties of coarse rice Pusa677 and Vallabh Bangani. Furthermore, N22 a non- Basmati variety was clustered with two traditional varieties Basmati-370 and Ranbir Basmati. The situation could be explained that the ISSR molecular markers could not represent DNA sequences of Basmati characteristics adequately. It also reflected inefficiency of ISSR markers. Govind and shahpasan were clustered together reflecting that pattern of clustering was independent of place of origin. The genotypes MAUB 13, MAUB 15 & MAUB 164 of quality rice developed at SVPUA&T Meerut were included in a single cluster, reason may be the common DNA sequences received from common parents. | |
| Conclusion | |
| The use of more number of markers would be efficient to characterize the three varieties i.e basmati, non-basmati & aerobic used for the present study, which highlighted the presence of diversity at genomic level among the genotypes studied. India harbors a huge resource of rice cultivars that are lesser known at the market front but hold great significance not only for farmers but also for the local consumers. An effort was made to collect a set of 30 cultivars including 10 basmati varieties, 10 non-basmati and 10 aerobic varieties of rice and to assess their genetic diversity. Their genetic diversity with molecular markers (ISSR RAPD markers) were according to their genotypes having far distant place of origin were clustered together reflecting that the distribution of varieties over clusters was independent of their place of origin/development like MAUB-57 the elite Basmati genotype could not be clustered with any variety. Also, NDR118 and Sathi varieties could not be grouped with other varieties. Some genotypes recently developed (Sathi, MAUB-13, MAUB-21 [Vallabh Basmati-21], MAUB- 15, MAUB-64 and MAUB-57) were also characterized and offered promise in their use in the genetic improvement of rice cultivars for grain quality even in case of SSR, out of 24 amplified bands, only 1 was monomorphic and 23 were polymorphic. In the RAPD assay, MAUB- 13, MAUB-15 and MAUB-64 were clustered with Taroari Basmati. Pusa Basmati-1 was clustered with Basmati-370. Another variety of quality rice PS-1121 was clustered with PS-2511. While MAUB-57 the elite Basmati genotype could not be clustered with any other variety. Also, NDR118 and Sathi varieties could not be grouped with other varieties. In SSR marker assay, all the Basmati varieties were clustered together. In addition, MAUB 13 and PS 2511, the non-Basmati genotypes were grouped in the same cluster. All varieties/genotypes adapted to aerobic conditions in Srilanka were also included in the same cluster along with Sarbati. Two genotypes shahpasan and Sathi were clustered together. ISSR+RAPD+SSR assay showed that all the genotypes of Basmati rice MAUB-13, MAUB-21 (Vallabh Basmati-21), MAUB-15, MAUB- 64 and MAUB-57 developed at SVPUA&T Meerut were clustered in cluster with Taroari Basmati. Pusa Basmati-1 was clustered with Basmati 370 and Ranbir Basmati. However, the most prominent variety of Basmati, PS 1121 was clustered with non- Basmati variety PS-2511 and could not be grouped in any cluster having traditional variety of Basmati. The dendogram also showed that Sathi and all genotypes of Srilanka origin well adapted to aerobic conditions were clustered together. | |
| The dendogram also showed that Sathi and all genotypes of Srilanka origin well adapted to aerobic conditions, which is a newly developed water-saving rice system in which rice grows in nonflooded and unsaturated soil according to 2001 report of the International Rice Research Institute in Philippines, this system has been monitored to identify potentially promising varieties of rice able to grow as an irrigated upland crop and quantify yield potential and water use efficiency, thus these aerobic varieties clustered together, which can be used for proper identification and selection of appropriate parents for breeding programs, including gene mapping, and ultimately for emphasizing the importance of marker-assisted selection (MAS) in aromatic/ non-aromatic rice improvement worldwide. | |
References
- value="1" id="Reference_Titile_Link">Sasaki T, Burr B (2000) International Rice Genome Sequencing Project: the effort to completely sequence the rice genome. Curr Opin Plant Biol 3: 138-141.
- value="2" id="Reference_Titile_Link">Onwueme IC, Sinha TO (1991) Field crop production in Tropical Africa, principles and practice CTA (Technical Centre for Agriculture and Rural Cooperation) Ede, The Netherlands. p p. 267-275.
- value="3" id="Reference_Titile_Link">Aggarwal RK, Shenoy V, Ramadevi J, Rajkumar R, Singh L (2002) Molecular characterization of some Indian Basmati and other elite rice genotypes using fluorescent – AFLP. Theor Appl Genet105:680-690.
- value="4" id="Reference_Titile_Link">Bhuiyan MAR (2005) Efficiency in evaluating salt tolerance in rice using phenotypic and marker assisted selection. M.Sc. dissertation, Department of Genetics and Plant Breeding, Bangladesh Agricultural University, Mymen singh, Bangladesh. pp.96.
- value="5" id="Reference_Titile_Link">Malik RK, Gupta RK, Singh CM, Brar SS, Singh SS, et al. (2008) Accelerating the adoption of resource conservation technologies for farm level impact on sustainability of rice–wheat system of the Indo-Gangetic plains, NATP Progress Report, CCSHAU, Haryana, India.
- value="6" id="Reference_Titile_Link">Hossain MM, Islam MM, Hossain H, Ali MS, Teixeira da Silva, et al.(2012) Genetic diversity analysis of aromatic landraces of rice (Oryza sativa L.) by microsatellite markers. Genes, Genomes and Genomics 6(SI1): 42-47.
- value="7" id="Reference_Titile_Link">Ravi M, Geethanjali S, Sameeyafarheen F, Maheswaran M (2003) Molecular marler based genetic diversity analysis in rice (Oryza sativa L.) using RAPD and SSR markers. Euphytica 133: 243-252.
- value="8" id="Reference_Titile_Link">Sivaranjani AKP, Pandey MK, Sudharshan I, Kumar GR, Madhav MS,et al.(2010) Assessment of genetic diversity among Basmati and non-Basmati aromatic rices of India using SSR markers. Current Sci 99: 221-226.
- value="9" id="Reference_Titile_Link">Kibria K, Nur F, Begum SN, Islam MM, Paul SK, et al. (2009) Molecular marker based genetic diversity analysis in aromatic rice genotypes using SSR and RAPD markers. Int J Sustain Crop Prod 4: 23-34.
- value="10" id="Reference_Titile_Link">Singh Y (2011) Molecular approaches to assess genetic divergence in rice.GERF Bulletin of Biosciences. 2: 41-48.
- value="11" id="Reference_Titile_Link">Chakraborty S, Vhora Z, Trivedi R, Ravikiran R, Sasidharan N (2013) Molecular studies of aromatic and non aromatic rice (oryza sativa l.) Genotypes for quality traits using microsatellite markers. The Bioscan 8: 359-362, 2013
- value="12" id="Reference_Titile_Link">Chakravarthi B K,Naravaneni R (2006) SSR marker based DNA fingerprinting and diversity study in rice (Oryza sativa. L). African J Biotech 5: 684-688.
- value="13" id="Reference_Titile_Link">Wei X, Yuan X, Yu H, Wang Y, Xu Q, et al. (2009) Temporal changes in SSR allelic diversity of major rice cultivars in China. J Genet Genomics 36: 363-370.
- value="14" id="Reference_Titile_Link">Rai SN, Rani V, Kojima T, Ogihara Y, Singh K P, et al. (2006) RAPD and ISSR fingerprints as useful genetic markers for analysis of genetic diversity, varietal identification, and phylogenetic relationships in peanut (Arachis hypogaea) cultivars and wild species. Genome 44: 763–772.
- value="15" id="Reference_Titile_Link">Banziger M, Long J (2000) The potential for increasing the iron and zinc density of maize through plant breeding.Food Nutr Bull 21:397–400.
- value="16" id="Reference_Titile_Link">Kaman Z, Kara B (2003)Genotypic variations for mineral content at different growth stages in wheat (Triticum aestivum L.). Cereal ResCommun 31: 459–466.
- value="17" id="Reference_Titile_Link">Nagaraju J, Kathirvel M, Kumar RR, Siddiq EA, Hasnain SE (2002) Genetic analysis of traditional and evolved Basmati and non-Basmati rice varieties by using fluorescence-based ISSR-PCR and SSR markers. Proc Natl Acad Sci U S A 99: 5836-5841.
- value="18" id="Reference_Titile_Link">FAO (2002) A report on: Crops and drops – making the best use of water for agriculture. Food and Agriculture Organization of the United Nations. Rome.
- value="19" id="Reference_Titile_Link">Weising K1, Atkinson RG, Gardner RC (1995) Genomic fingerprinting by microsatellite-primed PCR: a critical evaluation.PCR Methods Appl 4: 249-255.
- value="20" id="Reference_Titile_Link">Möller EM, Bahnweg G, Sandermann H, Geiger HH (1992) A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res 20: 6115-6116.
- value="21" id="Reference_Titile_Link">Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32: 314-331.
- value="22" id="Reference_Titile_Link">Provost and Wilkinson (1999) A new system of comparing PCR primer applied ISSR fingerprinting of potato accessions.Theor Apple Genet 98:107-117
- value="23" id="Reference_Titile_Link">Rohif FJ (2002) NTSYS-pc: Numerical taxonomy and multivariate analysis system (Ed. 2.2), Department of Ecology and evolution, State University of NY, Stony Brook
- value="24" id="Reference_Titile_Link">Saini N, Jain N, Jain S,Jain RK (2004) Assessment of genetic diversity within and among Basmati and non-Basmati rice varieties using AFLP, ISSR and SSR markers.Euphytica 140: 133-46.
- value="25" id="Reference_Titile_Link">Singh D, Kumar A, Sirohi A, Kumar R, Yadav R,et al. (2009) Effects of adaptation to irrigate and aerobic agro-eco systems and quality on ISSR and SSR based genetic analysis of Basmati and non-Basmati rice’s of Asia: Limitations of molecular tools. In: International Conference on “Current Trends in Biotechnology & Implications in Agriculture” held on 19-21 Feb. 2009 at SVBP University 0f Ag. &Technology, Meerut, Pp. 38-49.
- value="26" id="Reference_Titile_Link">Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74: 5463-5467.
- value="27" id="Reference_Titile_Link">Vikram P, Swamy BPM, Dixit S, Ahmed HU, Sta Cruz, et al (2012) Bulk Segregant Analysis: "An effective approach for mapping consistent-effect drought grain yield QTLs in rice" Field Crops Res. 134:185-192.
- value="28" id="Reference_Titile_Link">Ram SG, Thiruvengadam V, Vinod KK (2007) Genetic diversity among cultivars, landraces and wild relatives of rice as revealed by microsatellite markers. J Appl Genet 48: 337-345.
- value="29" id="Reference_Titile_Link">Upadhyay P, Singh V K, Neeraja C N (2011) Identification of Genotype Specific Alleles and Molecular Diversity Assessment of Popular Rice (Oryza sativa L.)Varieties of India. International Journal of Plant Breeding and Genetics 5: 130–140.
- value="30" id="Reference_Titile_Link">Neeraja CN, Vemireddy LR, Malathi S, Siddiq EA (2009) Identification of alternate dwarfing gene sources to widely used Dee-Gee-Woo-Gen allele of sd1 gene by molecular and biochemical assays in rice (Oryza sativa L.). Electronic Journal of Biotechnology
- value="31" id="Reference_Titile_Link">Narshimulu G, Jamaloddin M, Vemireddy LR, Anuradha G, Siddiq E (2011) Potentiality of evenly distributed hypervariable microsatellite markers in marker-assisted breeding of rice. Plant Breeding 130: 314–320.
- value="32" id="Reference_Titile_Link">Zhang SB, Zhu Z, Zhao L, Zhang YD, Chen T, et al. (2007) Identification of SSR markers closely linked to eui gene in rice. Yi Chuan 29: 365-370.
- value="33" id="Reference_Titile_Link">Ma H, Yin Y, Guo ZF, Cheng LJ, Zhang L, et al (2011) Establishment of DNA fingerprinting of Liaojing series of japonica rice. Middle-East Journal of Scientific research 8: 384-392.
- value="34" id="Reference_Titile_Link">Peng ST, Zhuang JY, Yan QC, Zheng KL (2003) SSR markers selection and purity detection of major hybrid rice combinations and their parents in China. Chin J Rice Sci 17: 15.
- value="35" id="Reference_Titile_Link">Zhou HF, Xie ZW, Ge S (2003) Microsatellite analysis of genetic diversity and population genetic structure of a wild rice (Oryza rufipogon Griff.) in China. Theor Appl Genet 107: 332-339.
- value="36" id="Reference_Titile_Link">Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20: 176-183.
Tables and Figures at a glance
| Table 1 | Table 2 |
Figures at a glance
 |
 |
 |
 |
||||
| Figure 1 | Figure 2 | Figure 3 | Figure 4 | ||||
 |
 |
 |
 |
||||
| Figure 5 | Figure 6 | Figure 7 | Figure 8 |
Relevant Topics
- Agricultural science
- Agronomy
- Climate impact on crops
- Crop Productivity
- Crop Sciences
- Crop Technology
- Field Crops Research
- Hybrid Seed Technology
- Irrigation Technology
- Organic Cover Crops
- Organic Crops
- Pest Management
- Plant Genetics
- Plant Breeding
- Plant Nutrition
- Seed Production
- Seed Science and Technology
- Soil Fertility
- Weed Control
Recommended Journals
Article Tools
Article Usage
- Total views: 16862
- [From(publication date):
May-2015 - Apr 03, 2025] - Breakdown by view type
- HTML page views : 12021
- PDF downloads : 4841
Peer Reviewed Journals
Make the best use of Scientific Research and information from our 700 + peer reviewed, Open Access Journals
