Commentary Open Access
Dissecting Signaling Pathways by the Use of Genetically-Encoded Biosensors: Dynamic Matters
Pauline Vandame1, Davel Trinel2, Jean-François Bodart1* and Corentin Spriet2
1University Lille, Régulation des Signaux de Division Team, UMR 8576 CNRS, F-59000 Lille, France
2University Lille, TISBio, UMR 8576 CNRS, F-59000 Lille, France
- *Corresponding Author:
- Jean-François Bodart
University Lille, Régulation des Signaux de Division Team
UMR 8576 CNRS
F-59000 Lille, France
Tel: 330320436867
E-mail: Jean-Francois.Bodart@univ-lille1.fr
Received date: February 2, 2016; Accepted date: May 12, 2016; Published date: May 18, 2016
Citation: Vandame P, Trinel D, Bodart JF, Spriet C (2016) Dissecting Signaling Pathways by the Use of Genetically-Encoded Biosensors: Dynamic Matters. Biosens J 5: 1000138. doi:10.4172/2090-4967.1000138
Copyright: © 2016 Vandame P, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Biosensors Journal
Abstract
Among biosensors, genetically-encoded FRET biosensors raised hope to dissect signaling pathways by monitoring enzymatic activities or second messengers levels in living cells and even in living and developing organisms. FRET (Froster resonance Energy Transfer) is a radiationless energy transfer from donor to acceptor fluorophore. For most genetically encoded donor/acceptor pair, this transfer can only occur if they are separated by a distance less than ~10 nm. A FRET based sensor is made up of an adapted bioreceptor tagged on both ends with appropriate fluorophores.
Keywords
Genetically-encoded biosensor; FRET; PKA; cAMP; AKAR; Epac
Genetically-Encoded FRET Biosensors for Signaling Networks
Cellular functions require an accurate transduction of extracellular stimuli into functional responses via intracellular signaling networks of second messengers and enzymes. Untangling the complexity of the signaling network has become imperative for the understanding of cell functioning both in physiological and pathological contexts, and for therapeutically perspectives. It might be achieved by focusing on specific protein kinases or second messengers, which act as main effectors or contributors in many pathological contexts, like those which serve as converging hub for oncogenes. . Among the tools within the arsenal of available biosensors for these signaling networks, genetically-encoded FRET biosensors raised hope to dissect signaling pathways by monitoring enzymatic activities or second messengers levels in living cells and even in living and developing organisms [1]. FRET (Förster resonance Energy Transfer) is a radiationless energy transfer from donor to acceptor fluorophore [2]. For most genetically encoded donor/acceptor pair, this transfer can only occur if they are separated by a distance less than ~10nm. A FRET based sensor is made up of an adapted bioreceptor tagged on both ends with appropriate fluorophores. The bioreceptor alters its conformation upon either second messenger presence or kinase activity and results in a change in FRET efficiency [3]. This change will be measurable in various manners while it induces modifications of most light properties (fluorescence emission, lifetime, polarization [4]).
AKAR and Epac as Archetypes for Genetically-Encoded FRET Reporters
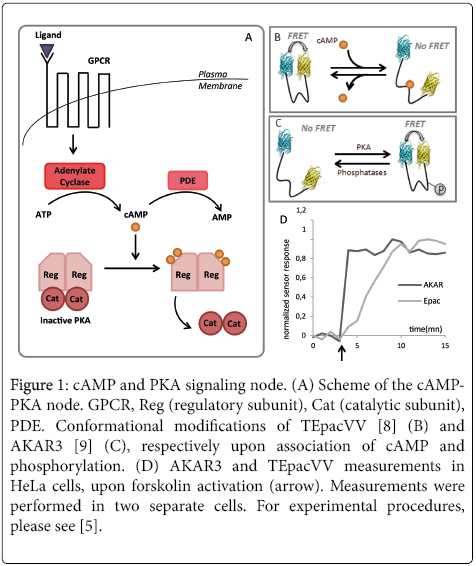
The cAMP-PKA (cyclic Adenosine monophosphate-a kinase protein) pathway is quite a node in molecular networks driving cellular responses towards external stimuli and exerts crucial functions throughout cell cycle [5]. In its inactive state, PKA is composed of regulatory subunits dimers associated with catalytic subunits dimer. Binding of a specific ligand to GPCR (G protein coupled receptor), leads to an increased production of cAMP by Adenylate Cyclase. The latter increase induces fixation of four cAMP molecules on the regulatory dimer resulting in the dissociation from the catalytic dimer, and represents the active form of the kinase. PKA activity can be down regulated by the degradation of cAMP achieved by the PDEs (PhosphoDiEsterase) (Figure 1A). The protein kinase A (PKA) activity, regulated by cAMP levels, has provided a focus of particular interest in the field of signaling pathways studies and a challenge for the engineering of genetically-encoded biosensors. To overcome the disadvantages of the biochemical methodologies, one had to cut a Gordian knot: how to extract individual response from a heterogenous cellular population, which might be at the same time transient, and sub-localized. Two families of genetically-encoded biosensors have emerged to this extent for PKA-cAMP tandem. PKA Kinase Activity Reporters (AKAR), are structurally composed of two fluorescent proteins, chosen for their amenability for FRET experiments, flanking a specific substrate for a specific kinase and a phospho-amino-acid binding domain (PAABD) [2]. This PAABD recognizes and binds the phosphorylated substrate, allowing a conformational modification that brings the fluorophores close to each other in order to lead to a measurable FRET signal. AKAR and its derivatives are the archetypes of KAR [6,7] and reflect the balance of phosphorylation/ dephosphorylation levels of PKA substrates. The other manner to get a hand on the pathway activity is to measure the level of the second messenger. Hence their name, cAMP sensors are based on the Epac proteins that are able to unfold upon cAMP binding. Thus in Epac sensor, the tandem of fluorophore is flanking a bioreceptor made of the Epac protein. In this case, the sensor is folded in absence of cAMP, while an increase of the cAMP level leads to a loss in FRET efficiency.
Figure 1: cAMP and PKA signaling node. (A) Scheme of the cAMPPKA node. GPCR, Reg (regulatory subunit), Cat (catalytic subunit), PDE. Conformational modifications of TEpacVV [8] (B) and AKAR3 [9] (C), respectively upon association of cAMP and phosphorylation. (D) AKAR3 and TEpacVV measurements in HeLa cells, upon forskolin activation (arrow). Measurements were performed in two separate cells. For experimental procedures, please see [5].
Different Viewing Angles on a Single Signaling Node
For both biosensors, one shall keep in mind that specificity of the phosphorylation/dephosphorylation site or cAMP association site are assessed usually through the use of pharmalogical compounds, whose specificities are always subjected to caution. In regard to their dynamic ranges and the choice of adequate controls, we also previously reported that genetically-encoded biosensors shall be carefully considered [7]. Furthermore, what is measured is the response of each specific biosensor to its environment (second messenger levels or kinase activity increases). Thus, the measured dynamic is a mix of both the biological pathway and the biosensor response time depending on its own structural properties. In regard to this matter, the tandem PKAcAMP is a school-case. Indeed cAMP fixation on Epac sensor will induce its opening while PKA activation will be reflected by bringing together the fluorophores into close vicinity. From a structural point of view, the two mechanisms are in complete opposition (Figure 1B and 1C), while both events are reflecting the activation of the signaling node involving cAMP-PKA. As dominoes, following the pathway activation, cAMP level first rises, and second, PKA activity is increased.
Practical Application of TEpacVV and AKAR3 and to Dissect cAMP/PKA Pathway
In this example, we used a well characterized acquisition and analysis procedure [5] to measure the previously described cascade in response to adenylate cyclase activation through forskolin. While one shall expect a maximum increase of CAMP level prior to PKA activation, we observed a response for AKAR3 that reaches its maximum within less than one minute, while TEpacVV reaches a plateau after five minutes, through a less abrupt slope that the one for AKAR3 (Figure 1D).
Though, without prior knowledge of the biological model, it could be hypothesized that PKA is activated by cAMP level less than TEpacVV detection threshold. We could also imagine that cAMP level required for PKA activation is quite low. This second hypothesis is also reinforced by the fact that, in this kind of experiments, high levels of drugs are used which produce very strong molecular responses. Nevertheless, it is more likely that the difference is the result of the intrinsic structural properties of the biosensors. Even if the tools used to monitor cAMP levels and PKA activity are both part of the genetically-encoded FRET-based biosensors family, they are two tools relying on different principles. It is, for example, possible that the unfolding of the cAMP sensor requires more time than the phosphorylation of AKAR3 by PKA. Indeed, in AKAR’s case, the kinase has to reach a free phosphorylation site within an unfolded sensor, whereas in Epac’s case, the cAMP molecules have to find their way in a folded sensor and thus schematically push the fluorophores away to access the binding site. We should neither forget about the dynamic ranges of sensors (that relies on fluorophores tandem among other parameters), resulting in variable FRET signal magnitude in response to the same pathways activation. While dissecting a signaling pathway with several genetically-encoded biosensors, it is thus worthy to assess the properties of relaxation-tightening of the biosensors by measuring the response time and amplitude to a massive activation of the pathway. Using two biosensors of different dynamical structures is tantalizing, in order to correlate the amount of second messenger and the activities of the kinases, but remains challenging.
Acknowledgements
PV was granted by the University of Lille 1 and the Region Nord- Pas-de-Calais. JFB is affiliated to the Site de Recherche Intégrée en Cancérologie (SIRIC ONCOLILLE). We thank the personal of the BICeL-Lille1-HB Facility for access to the microscopy systems and technical advices. We also thank, Jin Zhang and Kees Jalink (The Nederlands Kanker Instituut) as sources for material cited in this article. We are indebted to the Research Federation FRABio for providing the scientific and technical environment conducive to achieving this work.
References
- Kamioka Y (2012) Live imaging of protein kinase activities in transgenic mice expressing FRET biosensors. Cell Struct Funct 37: 65-73.
- Vandame P (2013) From FRET imaging to practical methodology for kinase activity sensing in living cells. Prog Mol Biol Transl Sci 113: 145-216.
- Li IT, Pham E, Truong K (2006) Protein biosensors based on the principle of fluorescence resonance energy transfer for monitoring cellular dynamics. Biotechnol Lett 28: 1971-1982.
- Jares-Erijman EA, Jovin TM (2003) FRET imaging. Nat Biotechnol 21: 1387-139
- Vandame P (2014) The spatio-temporal dynamics of PKA activity profile during mitosis and its correlation to chromosome segregation. Cell Cycle 13.
- Vandame P (2013) Optimization of ERK activity biosensors for both ratiometric and lifetime FRET measurements. Sensors (Basel) 14: 1140-115.
- Vandame P, Trinel D, Spriet C, Bodart J (2015) Genetically-encoded Biosensor for Kinases: To See is to Believe? Biosens J 4: 113.
- Klarenbeek JB, Goedhart J, HinkA, Gadella TW,Jalink KA (2011) mTurquoise-based cAMP sensor for both FLIM and ratiometric read-out has improved dynamic range. PLoS One 6: e19170.
- Allen MD, Zhang J (2006) Subcellular dynamics of protein kinase A activity visualized by FRET-based reporters. BiochemBiophys Res Commun 348: 716-721.
Relevant Topics
- Amperometric Biosensors
- Biomedical Sensor
- Bioreceptors
- Biosensors Application
- Biosensors Companies and Market Analysis
- Biotransducer
- Chemical Sensors
- Colorimetric Biosensors
- DNA Biosensors
- Electrochemical Biosensors
- Glucose Biosensors
- Graphene Biosensors
- Imaging Sensors
- Microbial Biosensors
- Nucleic Acid Interactions
- Optical Biosensor
- Piezo Electric Sensor
- Potentiometric Biosensors
- Surface Attachment of the Biological Elements
- Surface Plasmon Resonance
- Transducers
Recommended Journals
Article Tools
Article Usage
- Total views: 13270
- [From(publication date):
June-2016 - Apr 02, 2025] - Breakdown by view type
- HTML page views : 12347
- PDF downloads : 923