Research Article Open Access
Differential Responses in Germination, Growth and Genes Expression of Cu/Zn- and Fe-superoxide Dismutase of Barley Under Salinity Stress
Nidal Odat*Department of Biotechnology, Al Balqa Applied University, Jordan
- *Corresponding Author:
- Nidal Odat
Department of Biotechnology
Al Balqa Applied University
Al-Salt 19117
Jordan
Tel: +962 5 349 1111
E-mail: nidalodat@gmail.com
Received date: June 24, 2017; Accepted date: July 04, 2017; Published date: July 11, 2017
Citation: Odat N (2017) Differential Responses in Germination, Growth and Genes Expression of Cu/Zn- and Fe-superoxide Dismutase of Barley Under Salinity Stress. Adv Crop Sci Tech 5: 296. doi:10.4172/2329-8863.1000296
Copyright: © 2017 Odat N. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Advances in Crop Science and Technology
Abstract
Soil salinity limits crop productivity by affecting the growth, physiology, and expression of stress-responsive genes. To evaluate which varieties of cultivated barley from Jordan are salt tolerant, five cultivars of barley (Hordeum vulgare L.) of different varieties and morphotypes (i.e., two-and six-rowed barley) were evaluated in terms of their germination, growth traits, and gene expression of Cu/Zn- and Fe-SODs to three levels of salinity (100, 200 or 300 NaCl mM). Germination and root length were significantly affected by moderate and high levels of salinity (200 and 300 mM NaCl) mainly in the varieties Athroh, Mutah, Acsad176, Rum, and to lesser extent in Yarmouk variety, possibly as a consequence of osmotic stress and/or ionic toxicity. Analysis of quantitative real-time PCR (qRT-PCR) showed differential expressions of both Cu/Zn- and Fe-SOD genes between varieties and genotypes (i.e., the sixrowed barleys-Athroh, Acsad176, and Rum-and two-rowed barley-Yarmouk and Mutah). Moreover, both genes were up-regulated by salinity of 300 mM NaCl in the Athroh, Yarmouk, and Acsad176 varieties. Altogether, the result revealed that responses to salinity in all traits of germination, root growth, and gene expression were dependent on the variety and genotype of studied barley. Accordingly, these results have helped us to distinguish between salttolerant and salt-susceptible genotypes of cultivated barley of Jordan that shall be useful to local farmers and breeders of barley.
Keywords
Salt tolerance; Barley; Gene expression; Cu/Zn-SOD; Fe- SOD; Jordan
Introduction
Improving crop performance in saline soil is an overarching goal of any breeding program where knowledge of differential responses of crop varieties to salinity is important to screen for stress-tolerant genotypes [1-3]. Soil salinity is a devastating stress factor that limits crop productivity by affecting the growth, and biochemical regulation, i.e., up-and down-regulation, of stress-responsive genes [4,5]. When a plant is exposed to high salinity, osmotic stress can occur, lowering the soil water potential and reducing the amount of water available to a plant’s root system, ultimately inhibiting germination and reducing root growth [6]. Moreover, high salinity causes plant toxicity through the accumulation of toxic ions (Na+ and Cl-) that disrupt important metabolic and physiological changes [7].
In addition to its effects on germination and growth a plant, salinity induces overproduction of reactive oxygen species (ROS), including singlet oxygen (1O2), hydroxyl radicals (.OH), and hydrogen peroxide (H2O2), that can cause oxidative damage to macromolecules and cellular membrane [8]. Stress-tolerant plants have efficient scavenging systems including several anti-oxidative enzymes that prevent the accumulation of ROS [9]. An important example of these enzymes is the superoxide dismutase family (SOD; EC1.15.1.1) which protects plant from abiotic stresses by catalyzing the dismutation of superoxide into oxygen (O2) and hydrogen peroxide (H2O2), which is further degraded by other enzymes [10]. Different isomers of SODs, named based on their metal group cofactor, have been found in different cellular organelles of plants; copper zinc (Cu/Zn-SOD) is located in the cytoplasm and chloroplasts, and iron SOD (Fe-SOD) is located in the chloroplast [11,12]. Physiologically, many studies have indicated that changes in the SOD-enzymatic activities and some metabolites are reflected in an increase in SOD mRNA transcripts in response to stress factors [13]. Nonetheless, to the best of our knowledge, there is few if any, studies have examined the effect of NaCl on the levels SODmRNA transcripts in crop plants, in contrast to many studies that examined the effect of salinity on enzyme activities and metabolites [10,14-16].
The agroecosystem in Jordan is characterized by prevalence of both arid and semiarid areas with a fluctuating annual rainfall and high evapotranspiration that contributes to increased soil salinity [17]. Five different barley (Hordeum vulgare L.) varieties are currently cultivated in Jordan: Athroh, Rum, Acsad176 (six-rowed barley), and Yarmouk, and Mutah (two-rowed barley). Some of these cultivars are known for their response variation to drought [18], yet there is a limited knowledge of their performances under salinity stress. Accordingly, the main objective of this study was to elucidate the variation in salinity responses of the five varieties of barley as assessed by germination, growth, and the expression levels of the Cu/Zn-SOD and Fe-SOD.
Materials and Methods
Plant materials, stress treatment, growth and stomatal parameters
Uniformly sized seeds of five varieties of cultivated barley-Yarmouk, Acsad176, Athroh, Rum and Mutah-that have been developed and released in Jordan were used in this study. Initially, the seeds were obtained from the National Center for Agricultural Research and Extension (NCARE) of Jordan. The seeds were surface-sterilized with 70% (v/v) ethanol for 5 min followed by a 2% commercial bleach (sodium hypochlorite) treatment for 3-5 min, and rinsed thoroughly in sterile distilled water. After stratification for a few days at approximately 4°C, ten seeds from each variety were germinated on 12 × 12 cm petri dishes onto a wet filter paper (Whatman No 1, Whatman International Ltd., Kent, UK) at 25°C in the dark with or without salt treatment at four NaCl levels (0 mM NaCl as a control, 100 NaCl mM, 200 NaCl mM, 300 NaCl mM). These four levels of salinity were choosen as they have been shown to cause several growth and physiological responses to discriminate differences in crop cultivars [19-22]. After 4 days of treatment, the percentage of germination was determined after radicle emergence, and the root lengths of germinated seedlings were measured after 9 days of treatments. For gene expression analysis, leaves from the barley varieties were collected on the third day after treatment with a 300 mM NaCl solution, and were quickly frozen in liquid nitrogen at -20°C until RNA isolation.
RNA isolation and cDNA synthesis
Total RNA was extracted from frozen barley tissues using an IQeasyTM Plus plant RNA extraction mini kit (iNtRON Biotechnology, Korea). The concentrations of RNA in the samples were measured spectrophotometrically (260 nm/280 nm; Biochrom, Cambridge, UK). The first-strand cDNA was synthesized by mixing 2 μg of RNA with 4 μl of Prime Scriptáµ?á´¹ RT reagent (Takara, Japan), and the final volume of the mixture was adjusted to 20 μl with RNase-free water (0.1% (v/v); diethylpyrocarbonate-treated water). The samples were then placed in a thermocycler (Biometra, Germany) for 45 minutes at 37°C, followed by 15 seconds at 85°C, and finally at 4°C for approximately 5 minutes. Amplified samples were then diluted to 50 ng/μl with sterile RNasefree water and stored at -20°C for gene expression analysis by quantitative real-time PCR (qRT-PCR).
Quantitative real-time PCR (qRT- PCR)
Differential expression analysis of SOD genes (Cu/Zn-SOD and Fe- SOD) was studied using qRT-PCR. PCR reactions were performed in a total volume of 25 μl consisting of 10 μl of Kappa Syber Fast qPCR reagent (KAPA Biosystems, MA, USA), 50 ng/μl synthesized template cDNAs, and 10 μM aliquots of each primer of the studied genes (Table 1) at each. The cycling conditions were as follows: 2 min/95°C, 10 s/ 95°C, 25 s/57°C, 25 s/60°C, and a final extension step of 2 min/60°C using a CFX96 touch real-time PCR system (Bio-Rad) instrument. This experimental procedure was conducted at least three times starting from cDNA synthesis. The threshold cycle (Ct) values of the triplicate PCRs were averaged, and relative quantification of the transcript levels was analyzed using the comparative Ct method [23].
| Name | Accession number | Primer sequence 5′-3′ | Amplicon Length(bp) | Tm (°C) |
|---|---|---|---|---|
| Actin | AY145451.1 | CTCCATCATGAAGTGTGACGTG GACGACCTTGATCTTCATGCTG |
151 | 61.77 61.83 |
| Cu/Zn-SOD | HM537232.1 | GGTGACACGACTAATGGATGC GGAATCTGGCTATCGACAATGG |
164 | 61.50 61.40 |
| Fe-SOD | AK375983.1 | CTATCAACCCACTTGCTTTCGG CTGCTTTACAAGGGTCTGGATG |
144 | 62.04 62.28 |
Table 1: Names of studied genes, their accession numbers, primer sequences, amplicon length, and temperature degree used in their amplification.
Statistical analysis
Data analysis was performed with SPSS for Mac (v20, SPSS Inc., Chicago, IL, USA). The differential responses to salinity at all parameters estimated in this study were executed by one- and two-way analyses of variance (ANOVAs) and Duncan’s Multiple-Range test (DMRT). The values shown in the results section are represented as the arithmetic mean ± standard error (S.E.; n=3), with p value ≤ 0.05 regarded as a significant difference. Before the performing of data analysis, the normality was checked (Shapiro-Wilk test), and when normality and homogeneity of variance were violated, the logarithmic transform was taken for the raw data. To measure the level of gene expression of SODs, the actin gene transcript was used as an internal control to quantify the relative transcript level of each target gene in each sample [24].
Results and Discussion
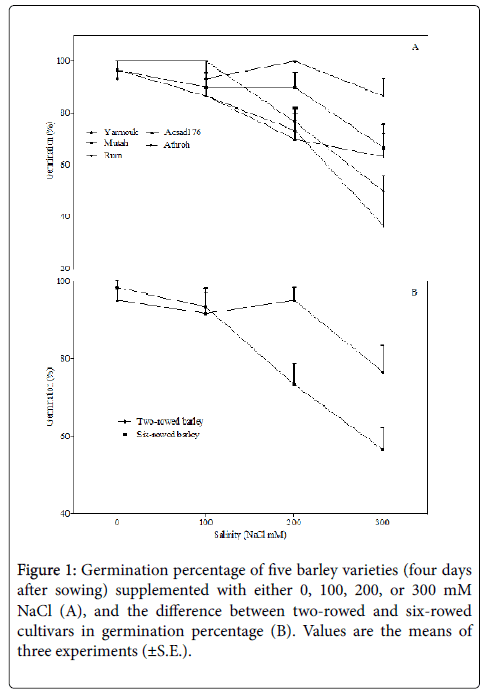
Although the differential responses of crop species and the varieties of a species to salinity is well documented, yet elucidating variations in crop responses of different genotypes of a crop plant to salinity is important as it helps in distinguishing plant varieties into tolerant and susceptible genotypes that are useful in breeding for stress tolerance [1]. Five varieties of barley from Jordan were evaluated for their germination, growth, and molecular responses to salinity. Under control conditions (0 mM NaCl), the germination percentage was similar in all barley varieties, and treating them with 100 mM NaCl did not significantly alter the germination. Contrarily, at a relatively moderate level of salinity (200 mM NaCl), the germination percentage was decreased significantly in the Athroh genotype (F1,4=49; p< 0.01), whereas the higher salt concentration (300 mM NaCl) was found to inhibit germination in the Acsad176 (F1,4=12.50; p< 0.05), Athroh (F1,4=75; p< 0.01), and Rum (F1,4=19; p< 0.05) (Figure 1A). Two-way ANOVA was used to examine the effect of salinity on germination in relation to the variation of cultivars, and the results showed that the salinity effect on germination is variety dependent (F12,40=1.9; p< 0.05, Table 2). Moreover, the inhibitory effect of 200 and 300 mM NaCl salinity on germination was found to be significantly stronger in six-rowed barley varieties (Athroh, Acsad176, and Rum) than in tworowed barley varieties (Yarmouk and Mutah) (Figure 1B). This inhibition of germination of all barley cultivars, except Yarmouk and Mutah, by salinity can be attributed to the water deficit caused by lower plant water uptake (known as osmotic stress) and by the cellular accumulation of toxic sodium and chloride ions that may interfere with the metabolic processes of germination [25,26]. However, it is also possible that both osmotic stress and ionic toxicity synergistically affected the germination process, and to distinguish them from each other, an investigation is required to compare the relative effects of NaCl and an iso-osmotic solution of an inert osmoticum such as polyethylene-glycol (PEG) [27].
| Source | Germination | Total root length |
|---|---|---|
| Salt treatment (T) | ** | * |
| Genotype (G) | ** | ** |
| T × G interaction | * | NS |
Table 2: Significance of the variation source (salt treatment, genotype, and treatment genotype interaction) in germination, total root length, and stomatal resistance of five varieties of barley under control or saline conditions.
As the germination of two-row barley varieties was found to be more tolerant to salinity than that of six-row varieties, it seems that genetic variation exists within the studied varieties that enabled some genotypes (two-rowed varieties) to germinate more efficiently than others under salinity stress. In fact, a recent study, based on a quantitative trait locus (QTL) study, suggested that two different regions in two different chromosomes of barley govern the germination capacity under varied levels of salinity [28]. This leads us to hypothesize that the studied genotypes of barley have different genetic potentials that, together with environmental conditions (i.e., salinity stress), can determine germination capacity.
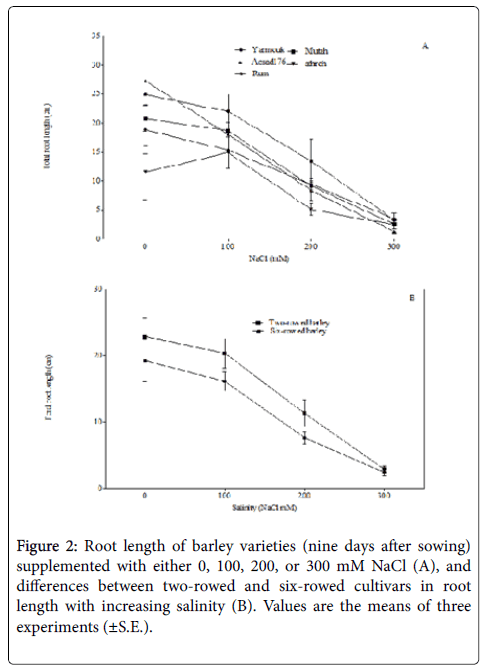
Root length was estimated in this study as an important determinant trait of successful seedling growth and development [29,30]. Root length in this study did not vary under normal conditions (i.e., 0 mM NaCl) and under salinity of 100 mM NaCl. Nonetheless, at a salinity of 200 mM NaCl root length was reduced significantly in accessions Mutah (F1,4=25.47; p< 0.01) and Acsad176 (F1,4=17.44; p< 0.05), while at 300 mM NaCl salinity, Yarmouk (F1,4=15.13, p < 0.05), Mutah (F1,4=57.77; p< 0.01), and Acsad176 (F1,4=38.24; p< 0.01) were found to show reduced root lengths (Figure 2A). Additionally, two-way ANOVA indicated that the salinity effect on root length is variety dependent and that a salinity of 200 mM NaCl decreased the root length of six-rowed genotypes more than it decreased the root length of two-rowed genotypes (F11,13=4.10; p=0.067 Figure 2B). The reduced root length caused by salinity may be due to the damage of seeds observed in ‘low-vigor seeds’, which is caused by the inhibition of cell division [4]. In addition, salinity can also reduce the water potential around the root system of a plant and thus decrease water and food availability to root cells [31].
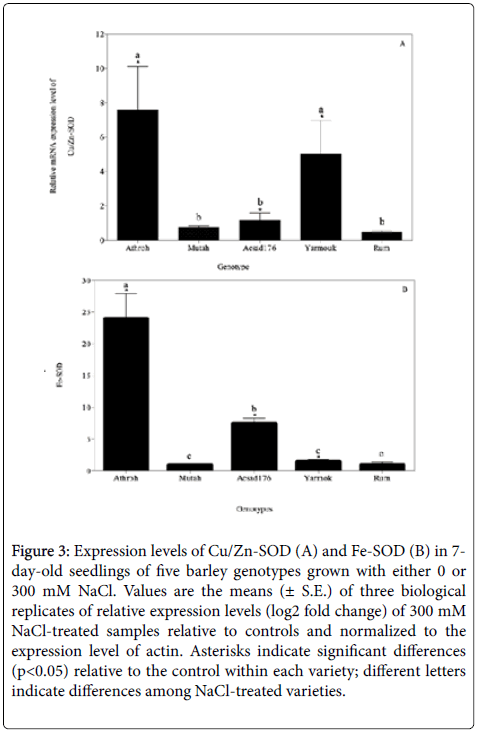
High salinity can also lead to the cellular accumulation of ROS that can damage certain macromolecules within a plant [8,32]. Such ROS are typically scavenged by a variety of anti-oxidative enzymatic families including superoxide dismutases (SOD) that are increased in activity in response to stress factors such as such salinity, drought, and pathogens [8,11,33,34]. The increased activity of these enzymes is typically indicative of the induction and enhancement of stressresponsive genes [12,35,36]. In this study, the expression levels (i.e., transcription) of the genes encoding anti-oxidative enzymatic isomers of SODs, namely, Cu/Zn-SOD and Fe-SOD, were studied in the barley cultivars using quantitative real-time PCR (qRT-PCR). The transcription levels of the genes were quantified in leaf tissues of genotypes exposed to 300 mM NaCl and compared to their corresponding untreated controls (0 mM NaCl). The results showed a differential expression of both Cu/Zn SOD (F4,10=5.76, p< 0.05), and Fe-SOD genes (F4,10=22.72, p< 0.001) in the varieties studied. Although, the Fe-SOD expression level was higher than the expression level of Cu/Zn-SOD, both genes showed a significant up-regulation in the Athroh, Yarmouk, and Acsad176 accessions upon exposure to salinity (Figure 3A and 3B). This up-regulation of both genes may be attributed to an increased generation of ROS by salinity that causes high activities of Cu/Zn-SOD and Fe-SOD enzymatic scavengers [36,37]. Moreover, the mean gene expression of Fe-SOD was higher in six-rowed barley than in two-rowed barley (mean of 10.37 and 1.19; F1,13=4.72, p< 0.05), suggesting that expression response of Fe/SOD is genotype dependent. The increased activity of SODs upon exposure to salinity has often been correlated to the degree of stress tolerance between plant species and between genotypes within species [14]. Accordingly, the gene expression analysis in this paper can lead us to the conclusion that Athroh, Yarmouk, and Acsad176 are salt-tolerant genotypes and that possibly possess an efficient ROS-scavenging system. The differential responses of these genes may suggest that they are good candidates for modulation of their enzymatic activities or for over-expressing them in different plant varieties in order to develop salt resistant genotypes [38]. Similar results of an increased induction of stress-related genes were found in stressed barley and wheat [5], maize [39], and cotton [40].
Figure 3: Expression levels of Cu/Zn-SOD (A) and Fe-SOD (B) in 7- day-old seedlings of five barley genotypes grown with either 0 or 300 mM NaCl. Values are the means (± S.E.) of three biological replicates of relative expression levels (log2 fold change) of 300 mM NaCl-treated samples relative to controls and normalized to the expression level of actin. Asterisks indicate significant differences (p< 0.05) relative to the control within each variety; different letters indicate differences among NaCl-treated varieties.
The dependence of the differential responses of germination, root length, and genes expression on variety and genotype of studied cultivars make them useful traits to distinguish the cultivated varieties into salt-tolerant and salt-susceptible genotypes [4,41,42]. Accordingly, the studied genotypes can be ranked as NaCl tolerant (number of traits indicated tolerance)>susceptible (number of traits indicated tolerance); Yarmouk (tolerant at 3 traits out of 3 total traits)>Acsad176 (2 traits)>Mutah ≈ Athroh ≈ Rum (1 trait). Therefore, Yarmouk seems to be the most salt-tolerant, possibly because they are able to adjust osmotically to high salinity levels, and Mutah, Rum, and Athroh are the most sensitive genotype.
Conclusion
This study provides knowledge on the performance of cultivated barleys from Jordan under differing salinity levels with respect to germination, growth, and gene expression. As the differential responses of most traits studied were found to depend on barley varieties and genotypes, it is, therefore, possible to distinguish the cultivated barley of Jordan into salt tolerant and salt susceptible genotypes that shall be useful to the local farmers and breeders of barley.
Acknowledgments
This study was funded by the deanship of scientific research of Al Balqa Applied University of Jordan. The author is grateful to Dr. Issam Qrunfleh and Dr. Yahaya Al-Shakhatreh for logistic help, Ms. Ruba Abu-Jamous and Ms. Ayah Awad for their excellent technical assistance, and the National Center for Agriculture Research and Extension (NCARE) of Jordan for providing barley seeds.
Conflict of Interest
The author declares no conflict of interest.
References
- Roy SJ, Negrao S, Tester M (2014) Salt resistant crop plants. CurrOpinBiotechnol 26: 115-124.
- Flowers TJ (2004) Improving crop salt tolerance. J Exp Bot 55: 307-319.
- Flowers TJ, Colmer TD (2008) Salinity tolerance in halophytes. New Phytol 179: 945-963.
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651-681.
- Temel A, Gozukirmizi N (2015) Physiological and Molecular Changes in Barley and Wheat under Salinity. ApplBiochemBiotechnol 175: 2950-2960.
- Flowers TJ, Hajibagheri MA (2001) Salinity tolerance in Hordeumvulgare: ion concentrations in root cells of cultivars differing in salt tolerance. Plant Soil 231: 1-9.
- Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25: 239-250.
- Miller GA, Suzuki N, Ciftci-Yilmaz S, Mittler RON (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33: 453-467.
- Ahmad P, Jaleel CA, Salem M A, Nabi G, Sharma S (2010) Roles of enzymatic and nonenzymatic antioxidants in plants during abiotic stress. Crit Rev Biotechnol 30: 161-175.
- de AzevedoNeto AD, Prisco JT, Enéas-Filho J, de Abreu CEB, Gomes-Filho E (2006) Effect of salt stress on antioxidative enzymes and lipid peroxidation in leaves and roots of salt-tolerant and salt-sensitive maize genotypes. Environ Exp Bot 56: 87-94.
- Kurepa J, Hérouart D, Van Montagu M, Inzé D (1997) Differential expression of CuZn- and Fe-superoxide dismutase genes of tobacco during development, oxidative stress, and hormonal treatments. Plant cell Physiol 38: 463-470.
- Alscher RG, Donahue, JL, Cramer CL (1997) Reactive oxygen species and antioxidants: relationships in green cells. Physiol Plant 100: 224-233.
- Rubio MC, Bustos-Sanmamed P, Clemente MR, Becana M (2009) Effects of salt stress on the expression of antioxidant genes and proteins in the model legume Lotus japonicus. New Phytol 181: 851-859.
- Sairam RK, Rao KV, Srivastava GC (2002) Differential response of wheat genotypes to long term salinity stress in relation to oxidative stress, antioxidant activity and osmolyte concentration. Plant Sci 163: 1037-1046.
- Brugnoli E, Lauteri M (1991) Effects of salinity on stomatal conductance, photosynthetic capacity, and carbon isotope discrimination of salt-tolerant (Gossypiumhirsutum L.) and salt-sensitive (Phaseolus vulgaris L.) C3 non-halophytes. Plant Physiol 95: 628-635.
- Sudhir P, Murthy SDS (2004) Effects of salt stress on basic processes of photosynthesis. Photosynthetica 42: 481-486.
- Szabolcs I (1989) Salt-affected soils. CRC Press, Inc.
- Samarah NH, Alqudah AM, Amayreh JA, McAndrews GM (2009) The effect of late-terminal drought stress on yield components of four barley cultivars. J Agron Crop Sci 195: 427-441.
- Rahnama A, Munns R, Poustini K, Watt MA (2011) A Screening method to identify genetic variation in root growth response to a salinity gradient. J Exp Bot 62: 69-77.
- Katsuhara M, Shibasaka M (2000) Cell Death and Growth Recovery of Barley after Transient Salt Stress. J Plant Res 113: 239-243.
- Hurkman WJ, Tanaka CK (1987) The effects of salt on the pattern of protein synthesis in barley roots. Plant Physiol 83: 517-524.
- Chen Q, Zhang WH, Liu YL (1999) Effect of NaCl, Glutathione and Ascorbic Acid on Function of Tonoplast Vesicles Isolated from Barley Leaves. J Plant Physiol 155: 685-690.
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3: 1101-1108.
- Livak KJ, Schmittgen TD (2001) Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-â??â??CT Method. Methods 25: 402-408.
- Mahajan S, Tuteja N (2005) Cold, Salinity and drought stresses: an overview. Arch BiochemBiophys 444: 139-158.
- Zhang H, Irving lJ, McGill C, Matthew C, Zhou Z, et al. (2010) The effects of salinity and osmotic stress on barley germination rate: sodium as an osmotic regulator. Ann Bot 106: 1027-1035.
- Dodd GL, Donovan LA (1999) Water potential and ionic effects on germination and seedling growth of two cold desert shrubs. Am J Bot 86: 1146-1153.
- Witzel K, Weidner A, Surabhi GK, Börner A, Mock HP (2009) Salt stress-induced alterations in the root proteome of barley genotypes with contrasting response towards salinity. J Exp Bot 60: 3545-3557.
- Hajlaoui H, El Ayeb N, Garrec JP, Denden M (2010) Differential effects of salt stress on osmotic adjustment and solutes allocation on the basis of root and leaf tissue senescence of two silage maize (Zea mays L.) varieties. Ind Crops Prod 31: 122-130.
- Shelden M, Roessner U, Sharp R, Tester M, Bacic A (2013) Genetic variation in the root growth response of barley genotypes to salinity stress. Funct Plant Biol 40: 516-530.
- Fogle VW, Munns DN (1973) Effect of salinity on the time course of wheat seedling growth. Plant Physiol 51: 987-988.
- Singh Gill S, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant PhysiolBiochem 48: 909-930.
- Mascher R, Nagy E, Lippmann B, Hörnlein S, Fischer S, et al. (2005) Improvement of tolerance to paraquat and drought in barley (Hordeumvulgare L.) by exogenous 2-aminoethanol: effects on superoxide dismutase activity and chloroplast ultrastructure. Plant Sci 168: 691-698.
- Lee SY, Ahn JH, Cha YS, Yun DW, Lee MC, et al. (2007) Mapping QTLs related to salinity tolerance of rice at the young seedling stage. Plant Breed 126: 43-46.
- Peiguo G, Michael B, Stefania G, Salvatore C, Guihua B, et al. (2009) Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. J Exp Bot 60: 3531-3544.
- Abu-Romman S, Shatnawi M (2011) Isolation and expression analysis of chloroplastic copper/zinc superoxide dismutase gene in barley. South African J Bot 77: 328-334.
- Murgia I, Tarantino D, Vannini C, Bracale M, Carravieri S, et al. (2004) Arabidopsis thaliana plants overexpressing thylakoidalascorbate peroxidase show increased resistance to Paraquat-induced photooxidative stress and to nitric oxide-induced cell death. Plant J 38: 940-953.
- Ghazi HB, Yamauchi Y, Shimada E, Sasaki R, Kawano N, et al. (2004) Enhanced tolerance to salt stress and water deficit by overexpressing superoxide dismutase in tobacco (Nicotianatabacum) chloroplasts. Plant Sci 166: 919-928.
- Pastori GM, Trippi VS (1993) Antioxidative protection in a drought-resistant maize strain during leaf senescence. Physiol Plant 87: 227-231.
- Gossett DR, Millhollon EP, Lucas M (1994) Antioxidant response to NaCl stress in salt-tolerant and salt-sensitive cultivars of cotton. Crop Sci 34: 706-714.
- Clipson NJW, Tomos AD, Flowers TJ, Jones RGW (1985) Salt tolerance in the halophyte Suaedamaritima L. Dum Planta 165: 392-396.
- Ashraf M (2009) Biotechnological approach of improving plant salt tolerance using antioxidants as markers. BiotechnolAdv 27: 84-93.
Relevant Topics
- Agricultural science
- Agronomy
- Climate impact on crops
- Crop Productivity
- Crop Sciences
- Crop Technology
- Field Crops Research
- Hybrid Seed Technology
- Irrigation Technology
- Organic Cover Crops
- Organic Crops
- Pest Management
- Plant Genetics
- Plant Breeding
- Plant Nutrition
- Seed Production
- Seed Science and Technology
- Soil Fertility
- Weed Control
Recommended Journals
Article Tools
Article Usage
- Total views: 3379
- [From(publication date):
August-2017 - Dec 22, 2024] - Breakdown by view type
- HTML page views : 2613
- PDF downloads : 766