Differential Association Between Responders HLA-DR Phenotypes and HLD-DR Antibody Production in Patients Awaiting for Renal Transplantation; Analysis of UNOS Database
Received: 20-Jan-2016 / Accepted Date: 20-May-2017 / Published Date: 27-May-2016 DOI: 10.4172/2475-7640.1000103
Abstract
Background: Immunogenicity of the Human Leukocytes Antigens (HLA) is highly variable. It is hypothesized that responder’s HLA-DR phenotypes contribute preferentially to produce antibodies against certain HLA-DR antigens. This study aims to stratify the immunogenicity of donor/responder’s HLA-DR phenotype combinations. Methods: Subjects studied were HLA-DR-homozygous patients waiting for kidney transplant in UNOS (United Network for Organ Sharing) database (n=2294) with anti-HLA-DR antibodies. Immunogenicity of recipient/donor DR combinations was determined by presence of significant positive and negative associations between HLA-DR phenotypes and HLA-DR antibodies as determined by likelihood analysis including odds ratios. Peptide binding affinity was determined with computer algorithms to corroborate our findings. Results: Out of 146 associations analyzed between HLA-DR phenotypes and antibody specificities, 88 combinations were significantly positive, 24 combinations were significantly negative, while 34 combinations were statistically insignificant. The highest positive association was seen between HLA-DR4-homogenous responders and anti-HLA-DR17 antibody (odds ratio= 4.05, p<0.0001). Directionality was found in the majority (68%) of acceptable mismatches. Some directionality in acceptability was explained by binding affinity between HLA-DR in responders and sensitizing indirect-pathway allopeptides. Conclusions: Generation of HLA-DR antibodies is influenced by recipient’s HLA-DR phenotypes. The results identified three groups of strong, irrelevant and acceptable HLA-DR mismatches. These findings support the possibility to predict, and therefore avoid, highly immunogenic donor-recipient HLA-DR combinations before kidney transplantation.
Keywords: Human leukocyte antigens (HLA); Antibodies; Renal transplantation
7552Abbreviations
AMR: Antibody-Mediated Rejection; CREG: Cross Reactive Group, DSA: Donor Specific Antibodies; HLA: Human Leukocytes Antigen; OR: Odds Ratio
Introduction
HLA antigens are the most polymorphic proteins in human. These antigens are very important for organ transplantation. It is well accepted that immunogenicity of the HLA mismatches is highly variable [1]. It has been recognized for long time that mismatched donor antigens are differentially recognised depending on the HLA phenotype of the recipient [2]. Analysis of clinical data from UNOS (United Network of Organ Sharing) demonstrated that the immunogenicity of all HLA class I was equal for kidney transplant [3]. Antibodies against donor HLA are associated with inferior graft function due to chronic rejection [4,5]. To avoid highly immunogenic mismatches, “taboo mismatches” stratagems were studied with either less mismatches on HLA epitopes [6-11] or matching within cross reactive group (CREG) [12-15]. HLDDR antigens have strong impact on graft survival [6].
Because donor-specific antibodies (DSA) to HLA-DR and DQ are found more frequently in post-transplant rejection [5], we were interested to identify high risk immunogenic HLA-DR mismatches as defined by significant association between recipient HLD-DR and alloreactive antibodies against certain HLA-DR antigens. Recently, sensitive solid-phase HLA antibody detection methods [16] were implemented in USA. Using large UNOS database, we aimed to determine odds of anti-HLA-DR antibody specificities associated with different HLA-DR phenotypes in responders.
Materials and Methods
We used data from the United States renal data System (USRD), a national data-reporting system that captures information on all American patients with end-stage renal disease on the waiting list for transplantation. Reporting to the USRD is mandatory for all centers that treat patients with end stage renal disease. This study was approved by the ethics review board of our institution. This study was restricted to HLA-DR-homozygous patients waiting for kidney transplant in recent UNOS (United Network for Organ Sharing) database during the 2012 year. Based on solid phase assays, antibody specificities were reported by UNOS histocompatibility laboratories. DR-homogeneity was defined on broad antigen level, i.e., patients with typing of DR13, 14, or DR6, 13, or DR6, 14 were all considered as DR6 homozygous. Broad (split) antigens in this study were: DR1 (DR1, DR103), DR2 (DR15, DR16), DR3 (DR17, DR18), DR5 (DR11, DR12), DR6 (DR13, DR14, DR1403, DR1404). HLA-DR-heterozygous patients were excluded from this study due to challenges to determine the relative contributions for each HLA-DR. Patients with negative panel reactive antibody (PRA) were excluded in this analysis as very likely they are not sensitized to HLA. HLA-DR-homozygous patients with antibody to HLA-DR (n=2294) were qualified for analysis.
Statistical analysis
Descriptive statistics were used to summarize the data. Chisquare test or Fisher’s exact test was used to compare the differences. Association strength between HLA-DR antibody specificities and HLA-DR phenotypes in responders was estimated by logistic regression analysis. Odds ratios (OR), 95% CI, and p-values were calculated. A 0.05 alpha level was used for statistical significance test. All analyses in this study were performed using SAS statistical software version 9.2 (The SAS Institute, Gary, NC).
Predict peptide binding abilities
On-line prediction tool IEDB (Immune Epitope Database and Analysis Program (http://tools.immuneepitope.org/main) [17] was used to predict peptide binding to HLA-DR. A consensus method was used based on 4 prediction algorithms: Neural network-based alignment (NN-align), stabilization matrix alignment (SMM-align), the combinatorial peptide scanning library and Sturniolo method. Median of percentile ranks was generated by comparing to 5 million random 15 mer peptides using Consensus method. SMM_align method was used for predicted IC50 (nM).
Results
The preferences of HLA-DR antibodies associate with responder’s HLA-DR phenotypes
As showed in Table 1, we had 2294 patients with homozygous HLA-DR antigens available for this study. The number of patients with homozygous HLA-DR10, and patients with antibodies to HLA-DR1403 or DR1404 were less than eight. Therefore, they were not analyzed. Subjects with listed antibodies to self HLA-DR were excluded in the analysis Nine recipient HLA-DR homozygous phenotypes were available and eighteen different HLA-DR antibodies were available for analysis. Out of 146 associations analyzed, 88 (60.3%) combinations between HLA-DR phenotypes and antibody specificity were significantly positive (OR>1, p<0.05). 24 (16.4%) combinations were found to have significant negative association (OR<1.0, p<0.05). 34 (23.3 %) combinations were statistically insignificant (p>0.05). These results indicate that immunogenicity is variable at the DR antigen mismatch level and that certain recipient/ donor HLA-DR combinations with heightened or diminished immunogenicity can be identified. For example, anti-HLA-DR1 antibodies were very likely produced in patients homozygous for HLA-DR3 (OR=1.92, p<0.0001), -DR5 (OR=1.89, p<0.0001), -DR6 (OR=1.87, p<0.0001), -DR7 (OR=2.83, p<0.0001), -DR8 (OR=2.3, p<0.0001) but not in patients homozygous for HLA-DR2 (OR=0.18, p<0.0001) and -DR4 (OR=0.4, p<0.0001). Patients homozygous for HLA-DR4 preferentially (p<0.05) produced antibodies to HLA-DR2 (DR15, 16), -DR3 (DR17, 18), -DR5 (DR11, 12), -DR6 (DR13, 14), -DR8, -DR9 and -DR52, but unlikely generate antibodies to HLADR1, -DR10 (p<0.05) and have no preference on making antibodies to HLA-DR7 and -DR51 (p>0.05). Notably, the most positive associations were anti-HLA-DR17 antibodies in patients homozygous for HLA-DR4 (OR=4.05, p<0.0001) or HLA-DR7 (OR=4, p<0.0001).
| Antibody | Responders with homozygous HLA-DRs | ||||||||
| To HLA-a | DR1 | DR2 | DR3 | DR4 | DR5 | DR6 | DR7 | DR8 | DR9 |
| % b | 5.93 | 15.9 | 13.4 | 17.9 | 11.5 | 19.3 | 10.3 | 4.01 | 1.44 |
| DR1 | NA c | 0.18 d | 1.92 d | 0.54 d | 1.89 d | 1.87 d | 2.83 d | 2.3 d | 0.81 |
| DR10 | 0.36 d | 0.43 d | 1.47 | 0.55 d | 1.32 | 1.94 d | 1.58 f | 1.73 | 0.54 |
| DR103 | 0.21d | 0.1 d | 2.25 d | 1.04 | 1.36 | 1.37 f | 2.23 d | 1.69 | 1.03 |
| DR11 | 2.01 d | 1.29 | 1.75 d | 2.37 d | 0.04 d | 0.45 d | 2.1 d | 0.44 e | 1.79 |
| DR12 | 1.51 | 1.33 | 1.43 f | 2.62 d | 0.46 d | 0.21 d | 1.82 d | 0.66 | 2.21 |
| DR13 | 2.56 d | 1.3 | 1.73 d | 3.14 d | 0.42 d | 0.04 d | 2.97 d | 0.45 e | 2.19 |
| DR14 | 1.8 e | 1.5 e | 0.83 | 1.92 d | 0.5 d | 0.28 d | 2.42 d | 0.67 | 1.51 |
| DR15 | 1.43 | NA | 1.49 f | 1.3 | 1.22 | 1.08 | 2.72 d | 1.89 f | 1.79 |
| DR16 | 1.34 | NA | 1.71 d | 1.36 f | 0.95 | 1.01 | 2.76 d | 1.95 f | 2.17 |
| DR17 | 2.95 d | 1.82 d | NA | 4.05 d | 0.51 d | 0.14 d | 4 d | 1.19 | 2.51 |
| DR18 | 2.57 d | 1.87 d | NA | 3.53 d | 0.37 d | 0.1 d | 3.19 d | 0.99 | 2.77f |
| DR4 | 1.69 f | 1.19 | 1.69 d | NA | 1.02 | 1.69 d | 2.48 d | 1.57 | 1.34 |
| DR51 | 0.53 | NA | 1.99 d | 1.04 | 1.5 f | 1.53 d | 1.77 d | 2.19 e | 0.77 |
| DR52 | 3.38 d | 3.41 d | NA | 2.65 d | NA | NA | 3.28 d | 2.2 e | 2.41 |
| DR53 | 0.95 | 1.34 | 1.96 d | NA | 2.2 d | 3.06 d | NA | 1.71 | NA |
| DR7 | 1.69 f | 1.8 d | 1.01 | 1.22 | 1.26 | 1.27 | NA | 1.13 | 1.34 |
| DR8 | 2.08 d | 1.23 | 1.26 | 2.33 d | 0.31 d | 0.3 d | 3.42 d | NA | 2.97f |
| DR9 | 1.22 | 1.33 f | 0.74 | 0.65 d | 1.3 | 1.34 f | 0.83 | 2.01 f | NA |
aThe number of patients with antibodies to HLA-DR1403 (N=24, 1%) and -DR1404 (N=24, 1%) are very low so the data are not included.
bPercentage of patients (N=2294) with homozygous HLA-DRs; The number of patients homozygous HLA-DR10 (N=8, 0.35%) are very low so their data are not shown.
cNA, the number is very low (N<8, 0.35%) so the data is not analyzed.
dP≤0.0001; e 0.0001<P≤0.001; f 0.001<p≤0.01; bold, p≤0.05; regular font, p>0.05 in Fisher’s two side test.
Table 1: Odds ratios for patients with homozygous HLA-DR to produce antibodies to a specific HLA-DR.
Graphic presentation of acceptable HLA-DR mismatches
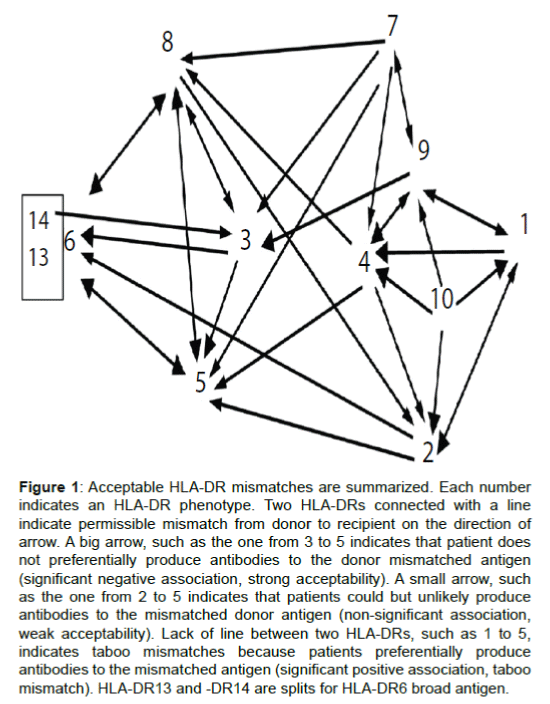
Based on Table 1 we presented Figure 1 to graphically display acceptable and unacceptable mismatches for better visibility. In this figure, split antigens with similar immunogenicity were combined as board antigens. Acceptable HLA-DR mismatches associated with decreased risk to generate antibodies are illustrated with arrows pointed from sensitizing to responder HLA-DRs. For example, HLA-DR5 homozygous patients were significantly less likely to produce antibodies to HLA-DR3, -DR6, -DR8 (Table 1). The big arrows from 3, 6 (13, 14), 8 to 5 in Figure 1 indicate strong acceptable mismatches. Small arrows from 2, 4, 7 to 5 indicate mismatches from HLA-DR2, -DR4 or -DR7 to HLA-DR5 were weakly acceptable because associations of antibodies to mismatched antigens and HLA-DR5 phenotypes in responders are not significant (p>0.05 in Table 1). Because HLA-DR5-homozygous patients preferentially produced antibodies to HLA-DR1, -DR9, -DR10 (Table 1), there was no links between points 1, 9, 10 to 5. This indicates mismatches from HLA-DR1, -DR9 or -DR10 to HLA-DR5 are at risk of generating antibodies and may be considered a taboo mismatches.
Figure 1: Acceptable HLA-DR mismatches are summarized. Each number indicates an HLA-DR phenotype. Two HLA-DRs connected with a line indicate permissible mismatch from donor to recipient on the direction of arrow. A big arrow, such as the one from 3 to 5 indicates that patient does not preferentially produce antibodies to the donor mismatched antigen (significant negative association, strong acceptability). A small arrow, such as the one from 2 to 5 indicates that patients could but unlikely produce antibodies to the mismatched donor antigen (non-significant association, weak acceptability). Lack of line between two HLA-DRs, such as 1 to 5, indicates taboo mismatches because patients preferentially produce antibodies to the mismatched antigen (significant positive association, taboo mismatch). HLA-DR13 and -DR14 are splits for HLA-DR6 broad antigen.
Importantly, we found that acceptability can be either bidirectional (for example, HLA-DR8 and -DR5) or unidirectional (for example, from HLA-DR3 to DR5) in other combinations. Surprisingly, the majority (17 out of 25, 68%) of acceptable HLA-DR mismatches were found to be unidirectional, rather than bidirectional (Figure 1). Notably, some of the strong acceptable mismatches were unidirectional, such as from HLA-DR3 to -DR5, from HLA-DR1 to HLA-DR4 and from HLA-DR9 to HLA-DR3.
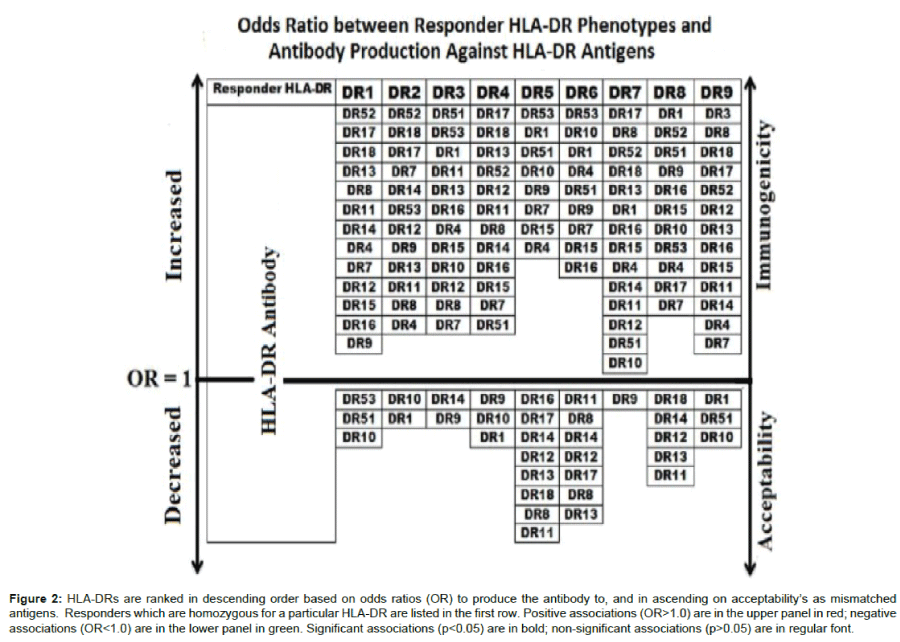
We then presented the odds ratio to recipient/ donor HLA-DR combinations in a graded scale in Figure 2. The arrow pointing upwards indicates an increasing likelihood of a recipient DR to associate with a given donor DR antibodies. The downward pointing arrow points to less immunogenic combinations with increasing Odds to have a significant negative association between donor DR and DR antibodies. For example, HLA-DR4-homozygous responders have highest odds to produce antibodies against HLA-DR17 followed by -DR18 and -DR13 and has the least odds to produce antibodies against HLA-DR1, followed by -DR10, and -DR9.
Figure 2: HLA-DRs are ranked in descending order based on odds ratios (OR) to produce the antibody to, and in ascending on acceptability’s as mismatched antigens. Responders which are homozygous for a particular HLA-DR are listed in the first row. Positive associations (OR>1.0) are in the upper panel in red; negative associations (OR<1.0) are in the lower panel in green. Significant associations (p<0.05) are in bold; non-significant associations (p>0.05) are in regular font.
The binding abilities of allopeptides with responders HLADR correlate with immunogenicity
To investigate why some HLA-DR mismatches are more immunogenic, we compared abilities of HLA-DR to indirectly present mismatched allopeptides. We postulate that help from indirect pathway CD4 T cells on humoral responses are intrinsically different, depending on both the types of HLA class II in responders and the nature of mismatched allopeptides. Binding ability of HLA-DRderived, 15-amino acid allopeptide to a specific HLA-DR binder was assessed with On-line prediction tool IEDB. Peptides with IC50 values (nM) <50, 500, 5000 are generally considered as high, intermediate, and low affinity respectively. Top percentile ranks, low IC50 indicate strong binding. As showed in Table 2, the strong binding affinity of sensitizing allopeptides with responder binders correlated with big odds of producing antibodies to sensitizing HLA-DR (Table 2). Especially, the strong binding for DRB1*13:01-allopeptides with HLA-DRB1*03:01 (Percentile Rank=1%, IC50=56nM) explains high likelihood of generating antibodies for HLA-DR13 to -DR3 mismatch (OR=1.7). While mismatches from HLA-DR3 to -DR6 (DR13 or DR14, OR=0.14,) or HLA-DR14 to -DR3 (OR=0.83) were not immunogenic due to weak binding of allopeptides with responder’s HLA-DR (Percentile Rank=19.5 ~ 66.3%). All mismatched amino acids except for one (26Y in DRB1*03:01 vs. 26F in DRB1*13:01/02/03) are included in the allopeptides (position 64-78) for prediction, so majority of indirect pathway alloreactivity in this mismatch are covered. Interestingly, a different allopeptide (position 3-17) derived from DRB1*03:01 was predicted to be a strong binder to either DRB1*04:01 (Percentile Rank=6.31, IC50=578), or DRB1*07:01 (Percentile Rank=5.91, IC50=319). This finding correlates well with strong preference of antibodies to HLA-DR17 (DRB1*03:01) in HLA-DR4 (OR=4.5) or -DR7 (OR=4.0) homogenous responders.
| DRB1* Proteina |
Allopeptide Sequence | DRB1* Binder d | % Rank e | IC50 (nM)f | Odds Ratio h |
| 13:01/2/3 | QKDILEDERAAVDTYb | 03:01 | 1 | 56 | 1.73 |
| 14:01 | QKDLLERRRAEVDTYb | 03:01 | 19.53 | 2947 | 0.83 |
| 03:01 | QKDLLEQKRGRVDNYb | 13:1/2/3 | 30.33 | 1737 | 0.14 |
| 03:01 | QKDLLEQKRGRVDNYb | 14:01 | 61.33 | NA | 0.14 |
| 03:01 | TRPRFLEYSTSECHFc | 04:01 | 6.31 | 578 | 4.05 |
| 03:01 | TRPRFLEYSTSECHFc | 07:01 | 5.91 | 319 | 4 |
aThe target sensitizing HLA from which the allopeptide is derived. The amino acid sequences are from position 64-78 b, or 3-17c. Letters highlighted in bold are mismatched amino acids comparing with DR binders.
dThe responders’ HLA used to predict binding ability for allopeptides.
eA top percentile (%) rank indicates high binding affinity of the peptide to the HLADR binder. Note for 13:01/02/03, average percentiles from each allele 13:01, 13:02, and 13:03 are used.
fPredicted with stabilization matrix alignment methods. Note for 13:01/2/3, average IC50 from each allele 13:01, 13:02, and 13:03 are used.
gNA: Prediction method is not available for this HLA allele.
hOdds Ratio for producing antibodies to sensitizing HLA in responders with phenotype of HLA-DR binders, adapted from Table 1.
Table 2: Association between HLA-Dr allopeptides and responder’s HLA-DRbinding affinity
Discussion
This is the first study to our knowledge that analyzed the relative immunogenicity of recipient HLA-DR and derived the likelihood and Odds ratios to generate HLA-DR Antibodies from the UNOS data set of patients waiting for transplantation with HLA-DR isotypes. It is known that the quantity [8] and physiochemical nature [18] of mismatched amino acids are critical for HLA immunogenicity. Here we added another parameter to this complexity. Phenotypes of the HLA-DR in the responder may contribute to humoral immunogenicity of HLA-DR. Our findings are consistent with previous studies regarding influence of responder’s HLADR phenotype on HLA class I immunogenicity [19-21].
Highly sensitive anti-HLA antibody screen and flow cytometry cross match methods have successfully reduced the risk of hyperacute rejection for organ transplant [22]. Nonetheless, patients with negative cross match and absent of DSA before transplantation may still develop de novo antibodies to donor mismatched HLA after transplantation [5] There is no decisive therapeutic treatment for rejection mediated by donor specific anti-HLA antibodies [5,23] The knowledge of immunogenicity of recipient/donor HLA combinations can be used to avoid immunogenic mismatches. A certain donor/recipient pair may be more likely to generate DSA and are at higher risk for antibodymediate rejection (AMR) than other pairs. For example in Figure 1, HLA-DR5-homozygous non-sensitized patients should receive organ transplant from donors carrying HLA-DR3, -DR6, -DR8, or to a less extent HLA-DR2, -DR4, -DR7, but not HLA-DR1, -DR9, -DR10. In Figure 2, the relative preferences for HLA-DR5-homozygous patients in donor HLA-DR mismatches are ranked from high to low according to odds to produce antibodies. This strategy can be used either for allocation of deceased donor organ or optimization in kidney paired donation. HLA-DR acceptability can be clustered in three groups: DR3- DR5-DR6-DR8, DR4-DR7-DR9, and DR1-DR2-DR10. The three broad HLA-DR groups are concordant with the known HLA-DR52-, DR53-, DR51- associated cross-reactive HLA-DRB1* groups. Anti-HLA-DR antibodies are generally easier to be produced inter-than intra-groups. If matching HLA-DR prolongs waiting time and disadvantage minorities, matching 3 broad HLA-DR groups may significantly reduce the likelihood to develop de novo DSA after transplantation.
A central finding in our study is that the majority of acceptable HLA-DR mismatches were uni-directional. Directionality was also found in some protective HLA class I and II mismatches based on cross reactivity [12] and in a unique combination of HLA-A28 to -A2 mismatch [24]. Notably, both we and the recent study [12] found the same uni-directional acceptability from HLA-DR1 to -DR4 mismatch. While we strongly believe HLA is the foundation for HLA immunogenicity, the directionality in immunogenicity can’t be solely explained with disparity in HLA amino acid between sensitizing and responder’s HLA-DRs. We hypothesize that HLA immunogenicity, particularly the directionality, depends on ability of HLA-class II in responders to indirectly present sensitizing allopeptides. In studies using small rodents, it was convincingly demonstrated indirect but not direct pathway CD4 T helper cells were able to help the isotype switching from IgM to IgG antibodies [25-27]. The different abilities for responder’s HLA-DR to bind indirect-pathway allopeptides may determine the strength of the indirect-pathway alloreactive CD4 T helper cells, which influence the ability to produce alloreactive antibodies to the mismatched HLA. This was true for some allopeptide bindings predicted with computer algorithm (Table 2). The allopeptide binding affinity also explained some directionality in acceptable mismatches, such as acceptable mismatches from HLA-DR3 to -DR6 while taboo mismatches from HLA-DR3 to -DR13 (an HLA-DR6 split antigen). Some of the binding affinity was confirmed with solid phase assays in vitro or HLA-transgenic mice in vivo (Zhou Q, Xu Q, unpublished data). Similarly, ability of class I allopeptide binding class II in responders has been found to correlate with the relative immunogenicity of HLAClass I, using either computer-based prediction algorithm [28-31] or in vitro binding assays [19,20]. Interestingly in a study that indirectly presented HLA-class I allopeptides correlated with de novo generated DSA in rejected kidney transplant. 68% of the allopeptides are not part of epitopes defined with the HLA-Matchmaker [31]. Involving responder’s HLA-DR may expend HLA humoral immunogenic into indirect alloresponses on T cell level, which are important both in chronic rejection [32] and in tolerance [33,34] of organ transplant.
There is clinical relevance to our work. Recent algorithms have been published to determine unacceptable HLA mismatches [35]. Considering HLA typing by serological methods remains widely in use internationally, this work can help clinicians to use this data to predict and possibly select recipient/donor acceptable HLA-DR mismatching combinations. Secondly the availability of Eplet matchmaker programs [36,37] is yet to be available for routine clinical use. Until such programs are routinely used, we recommend profiling the potential kidney transplant recipient based on panel reactive antibodies, donor specific antibodies and the potential immunogenicity reported in this study.
There are limitations in this study. Association does not mean cause and effect. Therefore further studies are needed to prove our conceptual framework. Absence of antibody to a specific HLA- DR may be explained by lack of previous immunization. Some strong negative associations between anti-HLA-DR antibodies and HLA-DR phenotypes in responder may be attributed to splits as partial selfantigens of broad antigens in responder’s phenotype, such as anti- HLA-DR11/12 antibodies in HLA-DR5 responders. Cut-offs and kits used to determine antibody specificities also vary among different UNOS HLA labs. The antibodies reported in UNOS might not always be real anti-HLA antibodies due to allo immunization, as there many ‘noise’ in solid phase antibody assay, such as naturally occurring anti- HLA antibodies, auto anti-HLA antibodies, antibodies to denatured antigens [38-41]. Immunogenicity of HLA mismatches were found to attribute to mismatched HLA epitope load [9-11] or cross-reactivity with self HLA [12]. A much bigger post-transplant cohort is necessary to determine contributions of recipient’s HLA-DR phenotype in immunogenicity of mismatched HLA.
Despite these limitations in immunization source, the relative immunogenicity for HLA-DR found in this study was not caused by random sensitization or overall hyperactivity in certain HLA-DR. Similarly to previous work [23] In spite of these limitations, this study identifies three broad categories with high, average and diminished risk to produce HLA-DR antibodies based on the recipient’s HLA-DR phenotypes. This knowledge can helps to preferentially select potential less immunogenic donor organs to avoid donor specific antibody generation.
Acknowledgments
We thank Saskatchewan Health Research Foundation for supporting this project.
Footnotes
Qingyong Xu participated in the design of the study, acquired and analyzed data, and wrote the initial drafts; H.J. Constructed and performed the statistical analysis of the data and reviewed the manuscripts; Ahmed Shoker participated in the design of study, interpretation of data analysis, and constructed the manuscript.
References
- Geneugelijk G, Thus KA, Spierings E (2014) PredicatingAlloreactivity in Transplantation. Journal of Immunology Research
- Doxiadis IN, Smits JMA, ThSchreuder GM, Presijn GG, Houwelingen HCV, et al. (1996) Association betweenSpecific HLA combinations and probability of kidney allograft loss: The taboo concept.The Lancet J 348: P850-P853.
- Sasaki N, Idica A, Terasaki (2008)Is there a differential strength of specific HLA mismatches in kidney transplants? Transplant Proc 40: 1091-1094
- Loupy A, Hill GS, Jordan SC (2012)The impact of donor-specific anti-HLA antibodies on late kidney allograft failure. Nat Rev Nephrol 8: 348-357.
- Wiebe C, Gibson IW, Blydt-Hansen TD, Karpinski M, Ho J, et al. (2012) Evolution and clinical pathologic correlations of de novo donor-specific HLA antibody post kidney transplant. Am J Transplant 12: 1157-1167
- Claas FH, Dankers MK, Oudshoorn M, van Rood JJ, Mulder A, et al. (2005) Differential immunogenicity of HLA mismatches in clinical transplantation. TransplImmunol 14: 187-191
- Duquesnoy RJ (2011) Antibody-reactive epitope determination with HLAMatchmaker and its clinical applications. Tissue Antigens 77: 525-534
- Duquesnoy RJ, Marrari M (2009)HLAMatchmaker-based definition of structural human leukocyte antigen epitopes detected by alloantibodies. CurrOpin Organ Transplant 14: 403-409.
- Sapir-Pichhadze R, Tinckam K, Quach K,Logan AG, Laupacis A, et al. (2015) HLA-DR and -DQ Eplet Mismatches and Transplant Glomerulopathy: A Nested Case-Control Study. Am J Transplant 15: 137-138
- Dankers MK, Witvliet MD, Roelen DL, de Lange P, Korfage, et al. (2004) The number of amino acid triplet differences between patient and donor is predictive for the antibody reactivity against mismatched human leukocyte antigens. Transplantation 77: 1236-1239.
- Wiebe C, Pochinco D, Blydt-Hansen TD, Ho J, Birk PE et al. (2013) Class II HLA epitope matching-A strategy to minimize de novo donor-specific antibody development and improve outcomes. Am J Transplant 13: 3114-3122.
- Lucas DP, Leffell MS, Zachary AA (2015) Differences in Immunogenicity of HLA Antigens and the Impact of Cross-Reactivity on the Humoral Response. Transplantation 99: 77-85.
- Takemoto S, Terasaki PI, Gjertson DW, Cecka JM (1994) Equitable allocation of HLA-compatible kidneys for local pools and minorities. N Engl J Med 331: 760-764.
- Sijpkens YW, Doxiadis, II, De Fijter JW, van Es LA, et al. (1999) Sharing cross-reactive groups of MHC class I improves long-term graft survival. Kidney Int 56: 1920-1927.
- McKenna RM, Lee KR, Gough JC, Jeffery JR, Grimm PC, et al. (1998) Matching for private or public HLA epitopes reduces acute rejection episodes and improves two-year renal allograft function. Transplantation 66: 38-43.
- Pei R, Lee JH, Shih NJ, Chen M, Terasaki PI (2003) Single human leukocyte antigen flow cytometry beads for accurate identification of human leukocyte antigen antibody specificities. Transplantation 75: 43-49
- Wang P, Sidney J, Kim Y, Sette A, Lund O, et al. (2010) Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics 11: 568.
- Kosmoliaptsis V, Dafforn TR, Chaudhry AN, Halsall DJ, Bradley JA, et al. (2011) High-resolution, three-dimensional modeling of human leukocyte antigen class I structure and surface electrostatic potential reveals the molecular basis for alloantibody binding epitopes. Hum Immunol 72: 1049-1059.
- Fuller TC, Fuller A (1999)Thehumoral immune response against an HLA class I allodeterminant correlates with the HLA-DR phenotype of the responder. Transplantation 68: 173-182.
- Papassavas AC,Barnardo MC, Bunce M, Welsh KI (2002)Is there MHC Class II restriction of the response to MHC Class I in transplant patients? Transplantation 73: 642-651.
- Dankers MK, Roelen DL, Nagelkerke NJ, de Lange P, Persijn GG et al. (2004) The HLA-DR phenotype of the responder is predictive of humoral response against HLA class I antigens. Human Immunology 65: 13-9.
- Zachary AA, Vega RM, Lucas DP, Leffell MS (2012) HLA antibody detection and characterization by solid phase immunoassays: methods and pitfalls. Methods MolBiol 882: 289-308.
- Archdeacon P, Chan M, Neuland C, Velidedeoglu E, Meyer J, et al. (2011) Summary of FDA antibody-mediated rejection workshop. Am J Transplant 11: 896-906.
- Dankers MK, Roelen DL, Van Der Meer-Prins EM, De Lange P, Korfage N, et al. (2003) Differential immunogenicity of HLA mismatches: HLA-A2 versus HLA-A28. Transplantation 75: 418-420.
- Sauve D, Baratin M, Leduc C, Bonin K, Daniel C (2004) Alloantibody production is regulated by CD4+ T cells' alloreactive pathway, rather than precursor frequency or Th1/Th2 differentiation. Am J Transplant 4: 1237-1245.
- Steele DJ, Laufer TM, Smiley ST, Ando Y, Grusby MJ,et al. (1996)Two levels of help for B cell alloantibody production. Journal of Experimental Medicine 183: 699-703.
- Taylor AL, Negus SL, Negus M, Bolton EM, Bradley JA, et al. (2007) Pathways of helper CD4 T cell allorecognition in generating alloantibody and CD8 T cell alloimmunity. Transplantation 83: 931-937.
- Papassavas AC, Stavropoulos-Giokas C (2002) Definition of the immunogenic HLA epitopes based on an epitope prediction algorithm. Transplantation Proceedings 34: 2049-2052.
- Papassavas AC, Stavropoulos-Giokas C, Boletis J, Iniotaki-Theodoraki JA, Ioannou S, et al. (2002) Epitope analysis of the HLA class I specific antibodies: a useful tool for the detection of the acceptable mismatches for highly sensitized patients. Transplantation Proceedings 34: 2053-2055.
- Papassavas AC, Stavropoulos-Giokas C, Boletis J, Ioannou S, Iniotaki-Theodoraki A, et al. (2002) Definition of permissible and immunogenic HLA antigens based on epitope analysis of the HLA specific antibodies produced in sensitized patients. European Journal of Immunogenetics 29: 401-407.
- Otten HG, Calis JJ, Kesmir C, van Zuilen AD, Spierings E (2013) Predicted indirectly recognizable HLA epitopes presented by HLA-DR correlate with the de novo development of donor-specific HLA IgG antibodies after kidney transplantation. Hum Immunol 74: 290-296.
- Liu Z, Sun YK, Xi YP, Maffei A, Reed E,et al. (1993) Contribution of direct and indirect recognition pathways to T cell alloreactivity. J Exp Med 177: 1643-1650.
- Xu Q, Lee J, Jankowska-Gan E, Schultz J, Roenneburg DA,et al. (2007) Human CD4+CD25low adaptive T regulatory cells suppress delayed-type hypersensitivity during transplant tolerance. J Immunol 178: 3983-3985.
- Xu Q, Lee J, Keller M, Burlingham WJ (2009) Analysis of indirect pathway CD4+ T cells in a patient with metastable tolerance to a kidney allograft Possible relevance to superior graft survival of HLA class II closely matched renal allografts. TransplImmunol20: 203-208.
- Süsal C, Roelen DL, Fischer G, Campos EF, Gerbase-DeLima M, et al. (2013) ClsasAlgorithms for determination ofunacceptable HLA antigen mismatches in kidney transplant recipients. TissueAntigens 82: 83-92.
- Silva E, Alba A, Castro A, Carrascal M, Buckel E, et al. (2010) Evaluationof HLA Matchcompatibility as predictorof graft survival and presence of ant-HLA antibodies Transplant Proc 42:266-269.
- Slavcev A (2013) Prediction of organ transplant rejection by HLA-specific and non-HLAantibodies--brief literature review. Int J Immunogenet 40: 83-87.
- Morales-Buenrostro LE, Terasaki PI, Marino-Vázquez LA, Lee JH, El-Awar N, et al. (2008) "Natural" human leukocyte antigen antibodies found in nonalloimmunizedhealthy males. Transplantation 86:1111-1115
- Ravindranath MH, Terasaki PI, Pham T, Jucaud V, Kawakita S (2013) Therapeutic preparations of IVIgcontainnaturally occurring anti-HLA-E antibodies that react with HLA-Ia (HLA-A/-B/-Cw) alleles. Blood 121:2013-2028
- Xu Q, Pearce T, Johnson E, Rich-Sperling D, Gorkoff K, et al. (2012) 32-OR: Are all anti-HLA Cw antibodies detected with luminex single antigen beads real antibodies. Human Immunology 73: 27
- Ravindranath MH, Terasaki PI, Maehara CY, Jucaud V, Kawakita S, et al. (2015) Immunoglobulin (Ig)G purified from human sera mirrors intravenous Ig human leucocyte antigen (HLA) reactivity and recognizes one's own HLA types, but may be masked by Fab complementarity-determining region peptide in the native sera. ClinExpImmunol 179:309-328
Citation: Shoker A, Xu Q, Lim HJ (2016) Differential Association Between Responder’s HLA-DR Phenotypes and HLD-DR Antibody Production in Patients Awaiting for Renal Transplantation; Analysis of UNOS Database. J Clin Exp Transplant 1: 103. DOI: 10.4172/2475-7640.1000103
Copyright: © 2016 Shoker A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 13929
- [From(publication date): 7-2016 - Nov 21, 2024]
- Breakdown by view type
- HTML page views: 13117
- PDF downloads: 812