Diagnostic and Prognostic Implications of AAGAB Expression in Human Breast Cancer
Received: 17-Oct-2022 / Manuscript No. DPO-22-77589 / Editor assigned: 20-Oct-2022 / PreQC No. DPO-22-77589 / Reviewed: 03-Nov-2022 / QC No. DPO-22-77589 / Revised: 10-Nov-2022 / Manuscript No. DPO-22-77589 / Published Date: 17-Nov-2022 DOI: 10.4172/ 2476-2024.7.S12.005
Abstract
Background: Alpha and Gamma Adaptin Binding protein p34 (AAGAB) was previously reported as a novel on- treatment biomarker can improve prediction of response to neoadjuvant chemotherapy in breast cancer. However, the expression and prognostic value of AAGAB in breast cancer is unknown, the function of AAGAB remains to be elucidated
Methods: Herein we investigated the role of AAGAB in human breast cancer from the GEO and TCGA databases, immunohistochemistry, Gene Set Enrichment Analysis (GSEA) and immune infiltration analysis
Results: The expression of AAGAB in breast cancer was significantly up-regulated relative to normal tissue (all p-values<0.05) in GEO and TCGA databases. Kaplan-Meier survival analysis showed that breast cancer patients with AAGAB-high had a worse prognosis than that with AAGAB-low (p=0.005). Univariate analysis using logistic regression revealed that age, pathological stage, and number of lymph nodes were significantly associated with poor Overall Survival (OS) (all p<0.05). Functional annotations indicated that AAGAB is involved in the most significant signaling pathways including intra Golgi traffic and peroxisomal lipid metabolism pathways.
Conclusions: Our study revealed that elevated AAGAB expression was significantly correlated with aggressive progression, poor survival in patients. AAGAB may serve as a new biomarker and potential treatment target in breast cancer.
Keywords: Breast cancer; AAGAB; Biomarker; Survival analysis; Prognosis
Abbreviations
AAGAB: Alpha and Gamma-Adaptin Binding Protein p34; OS: Overall Survival; IARC: International Agency for Research on Cancer; TCGA: The Cancer Genome Atlas; IHC: Immunohistochemistry; NES: Normalized Enrichment Score; GSEA: Gene Set Enrichment Analysis.
Introduction
Breast cancer is a malignant tumour occurring in mammary epithelial tissue, which proliferating out of control via the action of various carcinogenic factors. According to the latest data from the International Agency for Research on Cancer (IARC) survey in 2018, incidence (24.2%) and mortality (15.0%) of breast cancer occupy the top positions among women cancer worldwide [1]. Over the last few decades, the incidence of breast cancer has been increasing year by year in China. The implementation of neoadjuvant treatment and the popularization of screening can significantly reduce the mortality of breast cancer [2]. However, the mortality rate from breast cancer has not decreased significantly in China, especially in the vast rural areas [3]. So far, scientists have not found the exact cause of breast cancer, but many risk factors have found associated with breast cancer.
As a highly heterogeneous disease, patients’ response to treatment and prognosis are different even if the clinical stage and pathological grade are the same [4]. Biomarkers are now widely used for early cancer screening and prognosis assessment. Common biomarkers for breast cancer include Estrogen Receptor (ER), Progesterone Receptor (PR), and Human Epidermal Growth Factor Receptor-2 (Her2) [5]. Despite the advances in screening and diagnosis techniques in recent years, which have greatly improved the survival rate of breast cancer patients, it remains one of the major diseases with the highest female mortality rates [6]. At present, most biomarkers that predict prognosis of breast cancer lack specificity, thus, it is clinically important to discover new biomarkers to enhance prognosis and individualized treatment.
AAGAB encodes the alpha and gamma adaptin binding protein p34, which interacts with the gamma-adaptin and alpha-adaptin subunits of complexes involved in clathrin-coated vesicle trafficking [7, 8]. Previous studies have found that heterozygous loss mutations in AAGAB are associated with palmoplantar keratodermas and Punctate palmoplantar keratoderma type I, which based on hyperproliferation within the punctate lesions [9]. AAGAB maybe involving in endocytic recycling of growth factor receptors such as EGFR, which can result in increasing cell division [10]. There was no close correlation between AAGAB expression and tumor in previous studies. Bownes et al. found that AAGAB is a novel on-treatment biomarker can improve prediction of response to neoadjuvant chemotherapy in breast cancer [11].
However, the expression and functions of AAGAB in breast cancer are unknown, the role of AAGAB in breast cancer need to be further confirmed.
This study attempts to evaluate the diagnostic and prognostic value of AAGAB expression in human breast cancer. GSEA was performed to gain the biological pathways involved in breast cancer pathogenesis related to AAGAB. According to our informational analysis, we indicated that the significant diagnostic and prognostic value of AAGAB for breast cancer, AAGAB may serve as a new biomarker and potential treatment target for patients with breast cancer.
Materials and Methods
TCGA and GTEx databases
The Cancer Genome Atlas (TCGA), supervised by the National Cancer Institute and the American Human Genome Institute, utilizes high-throughput genomic analysis technology to analyze gene mutations, which have improved the ability to prevent, diagnose and treat cancer [12].
Immunohistochemistry (IHC)
Paraffin-embedded tumour sections of breast cancer (from Tongji Medical College of HUST) were incubated overnight at 4°C with rabbit anti-AAGAB polyclonal antibody (FLJ11506, from Novus) that was diluted 1:200 in TBST containing 1% NGS. After washing, the sections were conjugated with goat anti-rabbit Horseradish Peroxidase (HRP) antibody (from Novus) at 1:5000 dilution for 30 min at room temperature, Subsequently, the sections were stained with 3,3-Diaminobenzidine (DAB) and counterstained with hematoxylin, as well as dehydrated through a graded ethanol series and sealed with neutral gum. Finally, the sections were observed using a light microscope (Olympus).
Immune infiltration analysis
To examine the interactions between tumor infiltrating immune cells and the expression of AAGAB, we performed integrated repository tool for tumor-immune system interactions (TIMER, http://cistrome. shinyapps.io/timer/), which contains 10,897 samples from diverse cancer types available in the TCGA database [13, 14].
Gene set enrichment analysis
GSEA is a computational method to determine whether an a priori defined gene sets is significant between two groups [15, 16]. We used MSigDB database C2 collection to perform GSEA. For the purpose of analysis, we divided breast cancer patients into AAGAB high group and AAGAB low group based on the expression level of AAGAB. Gene set permutations were 1000 times for each evaluation. The nominal p value and Normalized Enrichment Score (NES) were used to sort the enriched pathways of gene sets.
Statistical analysis
All statistical analyses were performed using R. The association between clinical pathologic features and AAGAB levels were analysed with the Wilcoxon signed-rank test and logistic regression. To further assess the prognostic significance of AAGAB in breast cancer, Kaplan-Meier analysis and multivariate Cox regression analysis were performed. The cut-off value of AAGAB expression was determined by its median value.
Results
The expression of AAGAB in breast cancer and its significant up-regulation
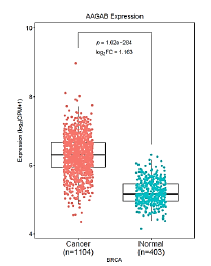
We screened the difference in AAGAB gene expression in the relevant cancer datasets from GEO and TCGA databases. The results demonstrated that the expression of AAGAB in GSE10780_GPL570, GSE42568_GPL570, GSE70947_GPL570 databases was significantly higher than that in normal tissues (P<0.05). 1104 breast cancer samples were included in this analysis and 403 normal breast tissues used for comparison. The expression of AAGAB in breast cancer was significantly up-regulated (p<0.001, log2FC=1.163) relative to normal tissue (Figures 1 and 2).
Figure 1: The expression profile of AAGAB in corresponding cancers from TCGA datasets. The cancer names marked in red represent AAGAB is significantly up-regulated, in blue represent AAGAB is significantly down- regulated, in black represent AAGAB is not significantly different. The difference threshold is |log2FC| ≥ 1, P-value ≤ 0.05. Note: ( ) Cancer; (
) Cancer; ( ) Normal.
) Normal.
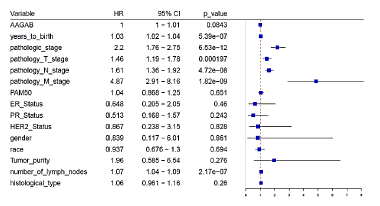
For the purpose of analyzing the association between clinicopathologic variables and AAGAB expression, we divided breast cancer patients into AAGABhigh group and AAGABlow group based on the expression of AAGAB, in which performed chi-square analysis. The pathological information of the patient includes: age, gender, ethnicity, pathological stage, TMN stage, PAM50 typing, typing, tumor purity, number of lymph nodes and histological type. The analysis shows that AAGAB expression was significantly associated with the patient’s age, gender, race, ER status, PR status, N-stage, PAM50 classification and histological type (p<0.05). For N-stage group, which is the case of lymph node metastasis, 43.39% (223/514) of patients of AAGAB high expression were in N0, 56.79% (205/361) of patients were in N1, 51.66% (62/120) of patients were in N2, 52.63% (40/76) of patients were in N3. The results indicate that the expression of AAGAB was relevant to the lymph node metastasis of patients. Regarding age, AAGABhigh group under 60 and over 60 was 43.82% (248/566) and 57.06% (291/510), respectively. These results indicate that the expression of AAGAB may be related to age, AAGAB expression also had a significant relationship with histological type, AAGAB was expressed low in medullary carcinoma and metaplastic carcinoma (Table 1).
| Characteristics | High expression (545) | Low expression (546) | X2 | p-value |
|---|---|---|---|---|
| Years to birth | 19.42105 | 6.06E-05 | ||
| <60 | 248 | 318 | ||
| ≥60 | 291 | 219 | ||
| Unknown | 6 | 9 | ||
| Pathologic stage | 4.639036 | 0.326378 | ||
| Stage i | 92 | 89 | ||
| Stage ii | 294 | 326 | ||
| Stage iii | 137 | 112 | ||
| Stage iv | 10 | 10 | ||
| Unknown | 12 | 9 | ||
| Pathology T stage | 8.566171 | 0.072907 | ||
| t1 | 145 | 134 | ||
| t2 | 308 | 323 | ||
| t3 | 63 | 75 | ||
| t4 | 28 | 12 | ||
| Unknown | 1 | 2 | ||
| Pathology N stage | 20.99004 | 0.000318 | ||
| n0 | 223 | 291 | ||
| n1 | 205 | 156 | ||
| n2 | 62 | 58 | ||
| n3 | 40 | 36 | ||
| Unknown | 15 | 5 | ||
| Pathology M stage | 0.340587 | 0.843417 | ||
| m0 | 455 | 453 | ||
| m1 | 12 | 10 | ||
| Unknown | 78 | 83 | ||
| gender | 4.141734 | 0.041838 | ||
| female | 535 | 544 | ||
| male | 10 | 2 | ||
| race | 31.7024 | 6.05E-07 | ||
| Asian | 35 | 26 | ||
| Black or African American | 61 | 121 | ||
| Unknown | 63 | 32 | ||
| white | 386 | 367 | ||
| PAM50 | 180.0719 | 7.20E-38 | ||
| Basal | 13 | 134 | ||
| Her2 | 26 | 41 | ||
| LumA | 268 | 158 | ||
| LumB | 133 | 51 | ||
| Unknown | 105 | 162 | ||
| ER Status | 23.54028 | 7.73E-06 | ||
| Negative | 4 | 32 | ||
| Positive | 39 | 29 | ||
| Unknown | 502 | 485 | ||
| PR Status | 16.4047 | 0.000274 | ||
| Negative | 12 | 39 | ||
| Positive | 32 | 22 | ||
| Unknown | 501 | 485 | ||
| HER2 Status | 3.412624 | 0.181534 | ||
| Negative | 32 | 45 | ||
| Positive | 11 | 16 | ||
| Unknown | 502 | 485 | ||
| Histological type | 23.71073 | 0.001281 | ||
| Infiltrating ductal carcinoma | 398 | 382 | ||
| Infiltrating lobular carcinoma | 94 | 109 | ||
| Medullary carcinoma | 0 | 6 | ||
| Metaplastic carcinoma | 0 | 9 | ||
| Mixed histology | 16 | 13 | ||
| Mucinous carcinoma | 13 | 4 | ||
| Other specify | 24 | 21 | ||
| Unknown | 0 | 2 |
Table 1: AAGAB expression associated with clinical pathological characteristics (Chi-square statistics).
Validation of elevated AAGAB protein level in breast cancer tissues
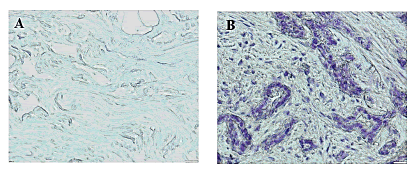
To assess AAGAB protein level in our paraffin-embedded tumor samples, we performed IHC staining and found that significant elevated AAGAB protein level in terms of density and intensity in breast cancer tissues compared to adjacent normal breast tissues (p=0.001) (Figure 3).
Survival analysis and multivariate analysis
In order to determine the effect of AAGAB gene expression levels on breast cancer prognosis, Cox multiple regression analysis was carried out. According to the expression of AAGAB, the cases were divided into the groups with the high AAGAB expression and the low AAGAB expression, followed by making the survival analysis. The results illustrated that there was no significant difference in disease- free survival between the high and low AAGAB expression groups, but a significant correlation was found for overall survival (p=0.005, indicated that breast cancer with AAGAB-high had a worse prognosis than that with AAGAB-low. Univariate analysis using logistic regression revealed that age, pathological stage, and number of lymph nodes were significantly associated with poor OS (all p<0.05) (Figures 4 and 5).
Figure 4: Disease-free survival curve and overall survival curve related to gene expression. There was no significant difference in disease-free survival between the high and low AAGAB expression groups (A), but a significant correlation was found for overall survival (p = 0.005, B). Note: A. ( ) Low expression (n=487); (
) Low expression (n=487); ( ) High expression (n=486); B. (
) High expression (n=486); B. ( ) Low expression (n=524); (
) Low expression (n=524); ( ) High expression (n=524)
) High expression (n=524)
Association of AAGAB’s expression with tumour purity and immune infiltration
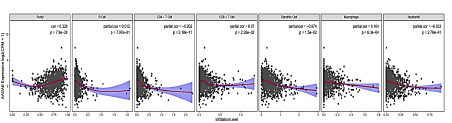
The above finding suggested that the expression of AAGAB may have an impact on the prognosis of breast cancer. The relationship between six types of immune infiltrating cells (including B cells, CD4 T cells, CD8 T cells, dendritic cells, macrophages and neutrophils) together with tumour purity and AAGAB expression was analysed to determine whether AAGAB expression was related to the level of immune invasion in cancer. AAGAB expression level was positively significantly correlated with cell purity (Cor=0.326, p=7.9E-28), CD8 T cells (Cor=0.07, p=2.26E-02), and macrophages (Cor=0.104, p=6.3E-04) of infiltration level. In addition, AAGAB expression level in breast cancer was negatively significantly correlated with CD4 T cells (Cor=-202, p=2.18E-11) and dendritic cells (Cor=-0.074, p=1.5E-02). Whereas, the expression of AAGAB in breast cancer was independent of B cells (Cor=0.012, p=7.07E-01) and neutrophils (Cor=-0.033, p=72.76E-01). In breast cancer, AAGAB expression levels were markedly positively correlated with tumour purity, indicating its relative enrichment in tumour cells. These findings indicate that AAGAB can affect the prognosis of breast cancer by interacting with breast cancer immune infiltration (Figure 6).
GSEA identifies differentially enriched AAGAB-related signaling pathways
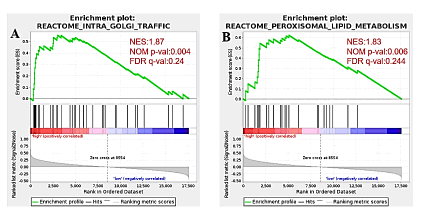
GSEA was conducted between high and low AAGAB expression gene sets to identify differentially enriched signaling pathways in breast cancer. Nom p-val<0.05, FDR q-val<0.25 are considered as significant. Two gene sets are significant at FDR<0.25 in phenotype high, 0 gene set is significant at FDR<0.25 in phenotype low. The Intra Golgi Traffic (p<0.004), Peroxisomal Lipid Metabolism (p<0.006) are differentially activated signaling pathways in AAGAB high expression phenotype (Figure 7).
Figure 7: Differentially enriched AAGAB-related signaling pathways identified by GSEA. Intra Golgi Traffic (p<0.004), Peroxisomal Lipid Metabolism (p<0.006) are differentially activated signaling pathways in AAGAB high expression phenotype. Note: ( ) Enrichment profile; (
) Enrichment profile; ( )Hits; (
)Hits; ( )Ranking metric scores
)Ranking metric scores
Discussion
Despite the advances in screening and diagnosis techniques in recent years, which have greatly improved the survival rate of breast cancer patients, it remains one of the major diseases with the highest female mortality rates. At present, most biomarkers that predict prognosis of breast cancer lack specificity, thus, it is clinically important to discover new biomarkers to enhance prognosis and individualized treatment.
AAGAB encodes the alpha- and gamma-adaptin binding protein p34, which interacts with the gamma-adaptin and alpha-adaptin subunits of complexes involved in clathrin-coated vesicle trafficking. Previous studies have found that heterozygous loss mutations in AAGAB are associated with punctate palmoplantar keratoderma, which based on hyperproliferation within the punctate lesions. AAGAB maybe involving in endocytic recycling of growth factor receptors such as EGFR, which can result in increasing cell division. The level of AAGAB was found to be prognostic of response in renal cancers and in thyroid cancers from the TCGA database. AAGAB was identified as a novel on-treatment biomarker for accurate prediction of pathological Complete Response (pCR) and reaction in patients treated with neoadjuvant chemotherapy. However, the exact role of AAGAB in breast cancer is currently unclear and warrants further investigation.
Herein we investigated the role of AAGAB in human breast cancer from the TCGA database, immunohistochemistry, GSEA and Immune infiltration analysis. Increased AABAB expression in breast cancer was significantly associated with age, gender, race, ER status, PR status, N-stage, PAM50 classification and histological type (all p-values<0.05). Kaplan-Meier survival analysis showed that breast cancer patients with AAGAB-high had a worse prognosis than that with AAGAB-low (p=0.005). Univariate analysis using logistic regression revealed that age, pathological stage, and number of lymph nodes were significantly associated with poor overall survival (OS) (all p<0.05). AAGAB expression level was significantly correlated with cell purity, CD8 T cells, macrophages, CD4 T cells and dendritic cells indicate that AAGAB can affect the prognosis of breast cancer by interacting with breast cancer immune infiltration.
Functional annotations indicated that AAGAB is involved in the most significant signaling pathways including intra Golgi traffic and peroxisomal lipid metabolism pathways. The underlying mechanisms between AAGAB and carcinogenesis of breast cancer remain unclear, elevated AAGAB maybe increase endocytic recycling of EGFR, which induce the signaling and division of breast cancer cells, however, future research is required to explore the detail mechanisms of AAGAB in breast cancer.
Conclusions
Our study revealed that elevated AAGAB expression was significantly correlated with aggressive progression, poor survival in breast cancer patients. AAGAB may serve as a new biomarker and potential treatment target in breast cancer. According to our informational analysis and IHC result, we prove the diagnostic and prognostic value of AAGAB in human breast cancer. GSEA was performed to gain the biological pathways involved in breast cancer pathogenesis related to AAGAB. AAGAB may serve as a new biomarker and potential treatment target for patients with breast cancer.
Compliance and Ethics
The study was approved by HUBU Ethics Committee. All patients have signed an informed consent.
Availability of Data and Materials
The datasets generated and analyzed during the current study are available in the https://xenabrowser.net/datapages/?hub=https://tcga. xenahubs.net:443 and https://www.gtexportal.org/home/.
Competing Interests
The authors declare that they have no competing interests.
Acknowledgements
We would like to thank Yuanyuan Zhao from Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology for her kindly help.
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, et al. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394-424.
[Crossref] [Google Scholar] [Pubmed]
- Munoz D, Near AM, van Ravesteyn NT, Lee SJ, Schechter CB, et al. (2014) Effects of screening and systemic adjuvant therapy on ER-specific US breast cancer mortality. J Natl Cancer Inst 106:289.
[Crossref] [Google Scholar] [Pubmed]
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, et al. (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66:115-132.
[Crossref] [Google Scholar] [Pubmed]
- von Wahlde MK, Timms KM, Chagpar A, Wali VB, Jiang T, et al. (2017) Intratumor heterogeneity of homologous recombination deficiency in primary breast cancer. Clin Cancer Res 23:1193-1199.
[Crossref] [Google Scholar] [Pubmed]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, et al. (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Nat Acad Sci U S A 98:10869-10874.
[Crossref] [Google Scholar] [Pubmed]
- Cardoso F, Harbeck N, Fallowfield L, Kyriakides S, Senkus E, et al. (2012) Locally recurrent or metastatic breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 23:11-19.
[Crossref] [Google Scholar] [Pubmed]
- Pohler E, Mamai O, Hirst J, Zamiri M, Horn H, et al. (2012) Haploinsufficiency for AAGAB causes clinically heterogeneous forms of punctate palmoplantar keratoderma. Nat Genet 44:1272-1276.
[Crossref] [Google Scholar] [Pubmed]
- Page LJ, Sowerby PJ, Lui WW, Robinson MS (1999) Gamma-synergin: an EH domain-containing protein that interacts with gamma-adaptin. J Cell Biol 146:993-1004.
[Crossref] [Google Scholar] [Pubmed]
- Giehl KA, Eckstein GN, Pasternack SM, Praetzel-Wunder S, Ruzicka T, et al. (2012) Nonsense mutations in AAGAB cause punctate palmoplantar keratoderma type Buschke-Fischer-Brauer. Am J Hum Genet 91:754-759.
[Crossref] [Google Scholar] [Pubmed]
- Rappoport JZ, Simon SM (2009) Endocytic trafficking of activated EGFR is AP-2 dependent and occurs through preformed clathrin spots. J Cell Sci 122:1301-1305.
[Crossref] [Google Scholar] [Pubmed]
- Bownes RJ, Turnbull AK, Martinez-Perez C, Cameron DA, Sims AH, et al. (2019) On-treatment biomarkers can improve prediction of response to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res 21:73.
- Tomczak K, Czerwinska P, Wiznerowicz M (2015) The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol 19:A68-A77.
[Crossref] [Google Scholar] [Pubmed]
- Li B, Severson E, Pignon JC, Zhao H, Li T, et al. (2016) Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 17:174.
[Crossref] [Google Scholar] [Pubmed]
- Li T, Fan J, Wang B, Traugh N, Chen Q, et al. (2017) TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 77:e108-e110.
[Crossref] [Google Scholar] [Pubmed]
- Yu G, Wang LG, Han Y, He QY (2012) Clusterprofiler: An R package for comparing biological themes among gene clusters. OMICS 16:284-287.
[Crossref] [Google Scholar] [Pubmed]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Nat Acad Sci U S A 102:15545-15550.
[Crossref] [Google Scholar] [Pubmed]
Citation: Liu Y, Zhang C, Cao W, Gong C, Liu Y (2022) Diagnostic and Prognostic Implications of AAGAB Expression in Human Breast Cancer. Diagnos Pathol Open S12 :005. DOI: 10.4172/ 2476-2024.7.S12.005
Copyright: © 2022 Liu Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Share This Article
Open Access Journals
Article Tools
Article Usage
- Total views: 2190
- [From(publication date): 0-2022 - Apr 06, 2025]
- Breakdown by view type
- HTML page views: 1789
- PDF downloads: 401


 ) Cancer; (
) Cancer; ( ) Normal.
) Normal.