Research Article Open Access
Development of Species-Specific Microsatellite Markers for Broomcorn Millet (Panicum miliaceum L.) via High-Throughput Sequencing
Min-Xuan Liu1, Yue Xu2, Tian-Yu Yang3, Zhi-Jun Qiao4, Rui-Yun Wang5, Yin-Yue Wang6and Ping Lu1*
1The National Key Facility for Crop Gene Resources and Genetic Improvement/Institute of Crop Science, Chinese Academy of Agricultural Science, Beijing 100081, China
2School of Life Science, Jilin University, Changchun 130012, China
3Institute of Crop, Gansu Academy of Agricultural Sciences, Lanzhou 030000, Gansu, China
4Institute of Crop Genetic Resources, Shanxi Academy of Agricultural Sciences, Taiyuan 030031, Shanxi, China
5Shanxi Agricultural University, Taiyuan 030031, Shanxi, China
6Faculty of Life Science, Jilin Agricultural University, Changchun 130118, Jilin, China
- *Corresponding Author:
- Ping Lu
The National Key Facility for Crop Gene Resources
and Genetic Improvement/Institute of Crop Science
Chinese Academy of Agricultural Science, Beijing 100081, China
Tel: +86-010-6215-9962
E-mail: lupingcaas@163.com
Received date: July 18, 2017; Accepted date: July 24, 2017; Published date: July 27, 2017
Citation: Liu MX, Xu Y, Yang TY, Qiao ZJ, Wang RY, et al. (2017) Development of Species-Specific Microsatellite Markers for Broomcorn Millet (Panicum miliaceum L.) via High-Throughput Sequencing. Adv Crop Sci Tech 5: 297. doi:10.4172/2329-8863.1000297
Copyright: © 2017 Liu MX, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Advances in Crop Science and Technology
Abstract
Objectives: To discover and develop large-scale SSR markers of the P. miliaceum genome, which can be used in future genetic studies effectively.
Result: 223,894 putative SSR sequences were identified by next-generation sequencing. A total of 56,694 primer pairs were successfully designed and 240 primer pairs were randomly selected for effectiveness validation. The expected heterozygosity and observed heterozygosity varied from 0.0447 to 0.7713 and from 0 to 0.9545, respectively and the mean of Shannon information index (I) was 0.7254. A UPGMA dendrogram indicated the high quality and effectiveness of these novel genomic SSR markers developed via next-generation sequencing technology.
Conclusion: A large repertoire of SSR markers were successfully developed by next-generation sequencing of the P. miliaceum genome which will be useful for the construction of genetic linkage maps, the identification of QTLs, and marker-assisted selection breeding.
Keywords
454 FLX titanium pyrosequencing; Marker development; Microsatellite; Panicum miliaceum L
Introduction
Broomcorn millet (Panicum miliaceum L, 2n=4x=36), an important member of the genus Panicum [1] was domesticated in China more than 10,000 year [2,3] and it is an outstanding crop for high adaptability to climate change, especially abiotic stresses, such as drought, salinity and infertility [4-6]. The growth duration of broomcorn millet is 60-90 days [7], which is the shortest in crops and it is usually grown for exploration of new wastelands and deserts or as a remediation crop in the event of natural disasters [8,9]. Although broomcorn millet grains are used primarily as bird and livestock feed in the United States and Europe the grains remain a staple for human consumption in China [10].
Over 8,600 accessions (varieties and landraces) of Panicum miliaceum have been deposited in the National Gene Bank located at the Chinese Academy of Agricultural Sciences (Beijing, China). Although abundant morphological variation exist among broomcorn millet landraces, the characterization and identification of this variation at molecular level is limited. This limitation is primarily due to the tetraploid genome (2n=4x=36) of P. miliaceum and the paucity of sequencing data, which has limited molecular marker development [11]. To date, no more than seventy characterized SSR loci are available for broomcorn millet [12,13], and these loci have been validated in relatively few genetic backgrounds. Initially, Hu et al. used 46 simple sequence repeat (SSR) markers from rice, wheat, barley and oat to study the genetic diversity of 118 broomcorn millet landraces collected from various ecological areas in China [12]. Through the construction of a SSR-enriched library from broomcorn millet genomic DNA, Cho et al. developed and identified 25 polymorphic microsatellite markers to analyze the genetic diversity of 50 P. miliaceum accessions from Mongolia, India, the Republic of Korea, Russia, Italy, and Uzbekistan [13].
Microsatellite is a type of DNA marker that is frequently used in many areas of research [14]. However, for the species which no genomic resources are available, the effective utilization and de novo isolation of SSR markers are limited [15]. The application of nextgeneration sequencing (NGS) technology has brought about a revolution in biological and agricultural applications because they can sequence DNA at unprecedented speed [16,17]. In conjunction with selective hybridization, NGS technologies can be used in highthroughput applications to develop and identify sequences that flank simple sequence repeat (SSR) regions. Species-specific SSR markers in mung beans [18], endangered dwarf bulrushes [19], fava beans [20], and grass peas [21] have been identified using this method and can be used as locus-specific markers to promote the study of downstream genotyping.
In this study, we used next-generation sequencing technology to inexpensively and efficiently obtain genomic SSR loci of broomcorn millet. Furthermore, 240 primer pairs were selected and amplified in 40 broomcorn millet genotypes aim to identify novel millet-specific SSR markers for future study.
Materials and Methods
Plant material and DNA isolation
Twenty-four broomcorn millet accessions with different seed color (white, grey, yellow, red, brown, and compound) were selected for 454 sequencing.
A set of 40 broomcorn millet accessions comprising 16 landraces and 24 cultivars from various geographic origins in China were used for SSR marker validation and genetic diversity analysis. Of these accessions, 5 were from Heilongjiang, 8 were from Shanxi, 12 were from Inner Mongolia, 4 were from Shaanxi, 5 were from Ningxia, 5 were from Gansu, and 1 was from Jilin.
Seeds were provided by the National Gene Bank of China located in Beijing, China. Detailed information of plant materials was listed in Table 1 (Additional file 1).
For SSR development, approximately equivalent weights of 7-dayold leaves (15-20) of each genotype were collected and pooled. Total genomic DNA was extracted using a cetyltrimethylammonium bromide (CTAB) method as modified by Edward et al. [22]. The concentration and purity of isolated DNA were determined using a NanoDrop ND-1000 (NanoDrop Technologies Inc., Wilmington, DE, USA).
Library preparation and 454 sequencing
Selective hybridization with streptavidin-coated beads with eight probes which including pAC, pGA, pAAG, pAAT, pAAC, pGATA, pATGT and pAAAT [23,24] was used to construct SSR-enriched libraries. Library quality was controlled by sequencing 192 clones which selected randomly from library. pEASY-T1 was used as the cloning vector, and insert fragments were validated by sequencing with an ABI3730XL DNA Analyzer. Libraries were considered high quality if sequence lengths were between 300 and 1,000 bp with a mean of 500-700 bp.
The eight DNA libraries enriched SSR probes were pooled in equal amounts, and then subjected to Roche 454 sequencing with GS-FLX Titanium system (Beijing Autolab Biotechnology Co., Ltd, China). The sequencing data was processed to generate a standard flow gram file (broomcorn millet.sff). This broomcorn millet.sff file was submitted to the SRA of National Center for Biotechnology Information (NCBI) with an accession number of SRX1223614.
Read characterization, SSR loci search and primer pair design
The sequencing data were pretreated with normalization, correction and quality-filtering algorithms, then processed to screen and filter out weak signals and low-quality reads. The read ends were trimmed using the EMBOSS software package for 454 adaptor sequences [25]. Furthermore, the length distribution of the reads and the nucleotide number in all reads was analyzed.
Before SSR loci search, the clean reads were filtered redundant by the CD-HIT program. A large-scale SSR search was done using the MISA tool. The minimum SSR motif length is 10 bp and monomer, dimer, trimer, tetramer, pentamer, and hexamer repeat lengths is 10, 6, 5, 5, 5, and 5, respectively. In a compound sequence, the max interruption between two SSR was set to 100 bp.
A Primer 3.0 interface module [26] was used for primer pairs design corresponding to the criteria proposed by Faircloth.
SSR marker validation and genetic diversity analysis
The characterizations including the total number of identified SSRs, the number of sequences containing more than 1 SSR, the number of SSRs present in compound formation etc. were analyzed using the MISA files [27] and plotted using R [28] and Open Office Calc.
Two hundred forty SSR primer pairs were selected and synthesized for polymorphic identification. Each PCR reaction (10 μL) containing 1.6 μL of 10 × PCR buffer (add Mg2+), 0.2 μL dNTP, 0. 5 U Taq DNA polymerase, 0.5 μL of a 5 μmol/L solution of each primer, 50 ng of gDNA templet, and 6.1 μL ddH2O. Primer pairs were amplified on a PTC-100 Thermo-Cycler (MJ Research, USA) with the following program: 5 min pre denaturation at 94°C; 39 cycles of 94°C for 45s, 50s annealing at 55°C, 1 min extension at 72°C; and a final extension at 72°C for 10 min. The PCR products were resolved using 8% polyacrylamide gel electrophoresis (PAGE). DNA bands were visualized by silver nitrate staining. Allele sizes were determined using a 50 bp DNA ladder (Dingguo, Beijing, China).
Through PCR amplification and PAGE testing, a total of 162 SSRs produced clear and reproducible polymorphic fragments and can be used in further study to assess the genetic diversity of the 40 P. miliaceum accessions from different geographical locations in China.
POPGENE1.31 [29] was used to assess genetic variability, including the observed total number of alleles (Na), effective allele number (Ne), Nei’1973 gene diversity (He) [30], and Shannon–Weaver index (I).
Genetic relationships between accessions were performed on the similarity matrix obtained from the proportion of shared fragments [31] using the NTSYS2.1 program and calculated using the unweighted Pair Group Method with Arithmetic Mean (UPGMA) cluster analysis.
Results
Quality evaluation of the SSR-enriched DNA library and read characterization of 454 sequencing data
The quality of the SSR enriched broomcorn millet library was tested by sequencing 192 randomly selected clones. The result showed that the recombination rate within the constructed P. miliaceum library was 86.5%, and 30.7% of the cloned sequence contained SSR motifs with an insert that varied between 200 and 1000 bp in size.
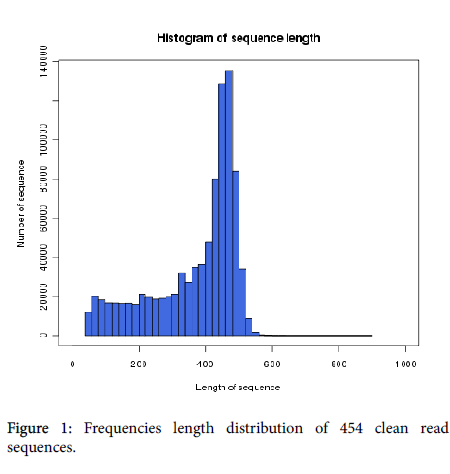
A total of 1,087,428 reads were generated using the Roche 454 GS FLX Titanium platform, and 904,311 reads were selected for next study after removal of adaptor. The most abundant nucleotide in the reads was adenine, accounting for 32.98% of the sequences, followed by cytosine (24.95%), guanine (23.71%), and thymine (18.34%). The average GC content was 48.66%. The most of read lengths were between 350 and 500 bp with a mean length of 370.4 bp and a maximum length of 565 bp (Figure 1).
Identification of SSR loci in the broomcorn millet genome
The microsatellite identification tool (abbreviated to MISA, and can be downloaded from (http://pgrc.ipk-gatersleben.de/misa/) was used for SSR loci mining. A total of 223,894 reads contained one SSR loci, and 289,155 SSRs were distinguished. Furthermore, there are altogether 45,604 sequences containing more than one SSR loci, and 61,908 containing compound SSR loci (Table 1).
| Category | Numbers |
|---|---|
| Total number of sequences examined | 904311 |
| Total size of examined sequences (bp) | 334957348 |
| Total number of identified SSRs | 289155 |
| Number of SSR containing sequences | 223894 |
| Number of sequences containing more than 1 SSR | 45604 |
| Number of SSRs present in compound formation | 61908 |
Table 1: MISA results from the genome survey.
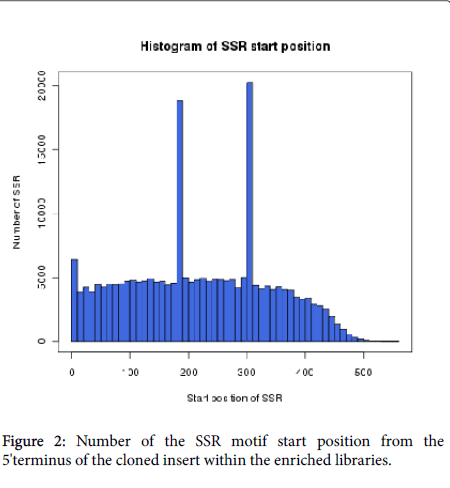
We analyzed the distribution of SSR loci start positions and found that a total of SSR motif reads length was 11,299,460 bp with an average valueof 199 bp. In the SSR motifs, most (78.6%) were situated within 320 bp of the 5’-terminus and middle regions of the cloned sequences. Few SSRs were located near the 3'-terminus (Figure 2). For later study of locus amplification, 56,694 SSR primers were successfully designed by the Primer 3.0 public shareware to meet the criteria including size range of amplification products, optimal melting temperature, GC content, etc (Additional file 2: Table 2).
| Primer pair ID | Repeat | F (5'-3') | R (5'-3') | Size (bp) | Ta(°C) |
|---|---|---|---|---|---|
| ICSBM2 | (GA)13 | GGCTTTGCTAGGGTTTCTCC | GGTGTGAAGTTGCCCAGATT | 226 | 60 |
| ICSBM3 | (GA)12 | GTGTCTCTTTCGTCTTGCCC | GGGACACTTCCACCATCATC | 204 | 60 |
| ICSBM5 | (GT)13 | TGTCTAGACCATCGCCATCA | CACTCACACACACATTTTCTTGG | 218 | 60 |
| ICSBM8 | (AC)14 | GTGGTACAGCTGCTCGTTCA | AGGAGGAACCAGGAAGCAAT | 254 | 60 |
| ICSBM10 | (AC)15 | GTGGTACAGCTGCTCGTTCA | GTGGTACAGCTGCTCGTTCA | 268 | 60 |
| ICSBM13 | (AC)16 | CGTTTTCTCGCTACACACGA | TGGACAACGGAAAACGTACA | 194 | 60 |
| ICSBM14 | (CA)11 | CTGCTGCATGCCTTTACCTT | CGCTGCAGTTTTGGTCAGTA | 252 | 60 |
| ICSBM15 | (CA)12 | ATGAATCACCCGATCCACAT | ACGCCAACATCAGCATATCA | 209 | 60 |
| ICSBM19 | (CA)13 | ATGAATCACCCGATCCACAT | ACGCCAACATCAGCATATCA | 211 | 60 |
| ICSBM21 | (CA)14 | GCTGTCGGTCAGTCCTGTTT | ACGCCAACATCAGCATATCA | 161 | 60 |
| ICSBM22 | (CT)12 | ACTCATGGTTACGGCAACTG | GCGCGAGAGAGAGAGAGAGA | 287 | 60 |
| ICSBM24 | (AC)25 | ATCGACGACTAGGCCCTGTA | GGCCGTCACTATATCTGTCACC | 153 | 60 |
| ICSBM27 | (CA)22 | CGATGAACGAAAATTCACCC | GTTCATTCGTCCAAATGCCT | 258 | 60 |
| ICSBM29 | (TG)16 | GAGATGGTGCGGATTCTGAG | TCATTTCCACTGTCACTGCC | 146 | 60 |
| ICSBM30 | (AC)18 | CAGAGCAGTGCGGTATTGTG | TCGTTTGTTGTTCGGTTGTC | 232 | 60 |
| ICSBM31 | (AC)21 | TCTGGACATGCTTTCACCAG | CCTACCTCGTAACACTGCGG | 267 | 60 |
| ICSBM33 | (GA)13 | AATATCCCTTTTGTCGCACG | ATGCATTGATGGGCTTGATT | 181 | 60 |
| ICSBM35 | (GA)10 | AGCAACGGAGGTGAGAGAGA | TCGACACACACGACACACAC | 128 | 60 |
| ICSBM39 | (CT)9 | TTTCAGGGACTGGACTGGAC | GTAGGGGGTAGCTGAGAGCC | 105 | 60 |
| ICSBM40 | (CT)8 | GCCTCCTGTCTTGTAGCGTC | AGGGTAGGCTGAGAGCCTGT | 121 | 60 |
| ICSBM43 | (CA)14 | GCACACGCATCATCACAAGT | GCTCATTCAACGACAGATGC | 280 | 60 |
| ICSBM46 | (GA)13 | CGTCCACCTTGGTGCTTATT | GCTGATTTTCTAACGGCTGC | 236 | 60 |
| ICSBM49 | (GA)19 | CTGCATTCTCTGTTCACCCA | ATCCTTTCACTCGAGGGGTT | 250 | 60 |
| ICSBM51 | (GT)17 | GCGCAGTAATATATTTCAGTAATTCA | GCATCATCGTCAAGACCTCA | 225 | 58 |
| ICSBM54 | (GT)18 | GCGCAGTAATATATTTCAGTAATTCA | GCATCATCGTCAAGACCTCA | 226 | 58 |
| ICSBM59 | (GT)19 | TCTTTTATGCGCGTAAGGCT | CACGAACACAAGAGAAGTAGCTCTCA | 262 | 59 |
| ICSBM60 | (AC)22 | ATCGACGACTAGGCCCTGTA | TGCGGAGTGTCTTTGTTCTG | 199 | 60 |
| ICSBM67 | (AC)23 | ATCGACGACTAGGCCCTGTA | TGTATGGAAAGCTCTGGCCT | 159 | 59 |
| ICSBM68 | (GT)10 | ATTTGACCTGTGACCTCGCT | AGGGCTCTCGAGGAGTGTTT | 195 | 60 |
| ICSBM71 | (GT)17 | GACCCAGCGATCAGTCTCTC | CTCTTGTCGTCTTGGTCCGT | 205 | 60 |
| ICSBM78 | (GT)21 | ACCCAACCGTATATCCAACG | TGTCACAGTTGTCCTGGCAT | 274 | 60 |
| ICSBM80 | (GT)9 | ATTTGACCTGTGACCTCGCT | CCTTTCTGTTTCTGCAAGCC | 215 | 59 |
| ICSBM81 | (TC)8 | CAACAAGGTTGGTTGGCTTT | ATGCTGCTGCAGATGTTTTG | 166 | 60 |
| ICSBM85 | (TG)15 | TGTGGGAGAGAAGTGGGC | CAAGGAAGGAATAAACCGCA | 187 | 59 |
| ICSBM86 | (AC)21 | AGTTAACCCTTGTGATGCCC | CGTTGTTGGTCCTTCTGGTT | 253 | 58 |
| ICSBM90 | (AC)8 | GCAGTGGGTCAGCTTATGGT | TCTCTCTGTGTGTGTGCGTG | 210 | 60 |
| ICSBM96 | (CA)16 | TGAGATTGGCATCAAGCAAG | TTTCTGGTCAGTTCGGTCAG | 287 | 58 |
| ICSBM99 | (CA)19 | GCCACACTAAATAAGCTTTGTGTC | TGGTCGTCACTGATTACGGA | 236 | 60 |
| ICSBM100 | (GA)15 | GAGTTAGAGGACAGCGTGGC | TGCAGCAGAGAATGTGCTACT | 210 | 58 |
| ICSBM107 | (GA)17 | GTCCTCACCTCCTTTTGGGT | CCTTCGTTTCTCTCTCGTCG | 250 | 60 |
| ICSBM109 | (GT)14 | TTCTCCGTCAGCTCACATTG | TCCATTGTTCATTTAGTAGAAACCT | 251 | 57 |
| ICSBM113 | (GT)15 | GACCCAGCGATCAGTCTCTC | CTCTTGTCGTCTTGGTCCGT | 201 | 60 |
| ICSBM119 | (GT)16 | GACCCAGCGATCAGTCTCTC | GACCTCACCTCTTCGTCGTC | 215 | 59 |
| ICSBM120 | (GT)22 | CGCACTAGCCCTTGTCTTTC | CGCCCTACGAACAAATCACT | 225 | 60 |
| ICSBM123 | (TAG)14 | CGAGTCGGTGAAGAGAGACC | TTTGCAATGTTCACCCAACT | 290 | 59 |
| ICSBM126 | (TC)8 | CAACAAGGTTGGTTGGCTTT | ATGCTGCTGCAGATGTTTTG | 165 | 60 |
| ICSBM127 | (AC)16 | TATTCGAGCCCCATTTCTTG | GCGTTATCCGGATGATGAAG | 184 | 60 |
| ICSBM130 | (AC)17 | CTGATCAAATCAATGCAGCAA | GTTTTTAGGTCCGTGGCGTAAAG | 132 | 60 |
| ICSBM132 | (CA)14 | CACACAGATATTTGGCACCG | TGAGGATCCGAAAAGATTGG | 216 | 60 |
| ICSBM135 | (CA)7 | GCCGGAGTATAGATCCGACA | GTCAGGCCGTGAACGTTATT | 175 | 60 |
| ICSBM139 | (CA)10 | ATGCACGCACGAACACATA | TCTTGATCATCACCAGCACC | 280 | 59 |
| ICSBM144 | (CA)8 | CACCATGTGTATGCGTGTGA | GGAGAGGAGCTTTCAGAACCA | 234 | 60 |
| ICSBM151 | (CA)10 | CCTCTCCTTACACGGGGATT | TTGATTATGCTTTGGAGGGG | 242 | 60 |
| ICSBM159 | (AC)13 | ATCGTAGAAACCATTGGCCC | TGACCCATGGACACTTTTCA | 279 | 60 |
| ICSBM165 | (AC)14 | AATGTCACAGGTTTCCCTCG | GCGAGAAAGAGGAGAGGGTT | 225 | 60 |
| ICSBM172 | (AC)6 | TATAGCCTCACCGCTCGTCT | GGCCTGAAAACTCAAATGGA | 206 | 60 |
| ICSBM180 | (CA)12 | TGGGACAATATGGCAAGGTT | ACAAATGCCTGATGGTAGGC | 237 | 60 |
| ICSBM197 | (CA)13 | CACACAGATATTTGGCACCG | TGAGGATCCGAAAAGATTGG | 214 | 60 |
| ICSBM205 | (CA)19 | ATTTTCTGGGCAATTCAACG | GTCCTCATCCCTTCCCTCTC | 191 | 60 |
| ICSBM206 | (CT)6 | GTGAAGAACTCTCGATCGGC | ACTGGGTAGTACACGGCGAG | 305 | 60 |
| ICSBM212 | (CT)6 | AGCACTGAGGCACAATTCCT | GTGCTGGGGTTTGTGACTTT | 231 | 60 |
| ICSBM216 | (GA)17 | CTACCGCTTCAAAACGAAGG | TGTCCCACTCTCCTACCTACTACC | 178 | 59 |
| ICSBM219 | (GA)20 | ATGGGATGCACAGGTACACA | TCCTTAGGTCATCGTCCTATTTG | 260 | 59 |
| ICSBM227 | (GT)11 | TTGATTATGCTTTGGAGGGG | CCTCTCCTTACACGGGGATT | 248 | 60 |
| ICSBM234 | (GT)15 | GACCCAGCGATCAGTCTCTC | TTGTCGTCTTGTCCGTCG | 198 | 59 |
| ICSBM235 | (TG)8 | GACCAGAGACTTGGGCTTTG | TCACTCACTCACTCATCCGC | 243 | 59 |
| ICSBM239 | (GA)6 | CCTGGACACACACACACACA | TCTTGTCACTGTCGGCGTAG | 233 | 60 |
Table 2: Characteristics of 66 polymorphic SSR markers developed in Panicum miliaceum L.
Abundance and length frequencies of SSR repeat motifs from broomcorn millet
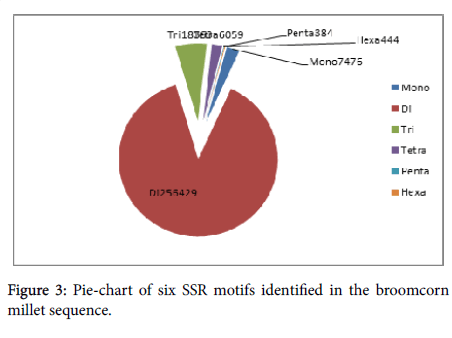
The total number of identified SSRs was 289,155, which included 7,476 mononucleotide repeat motifs (2.59%), 256,429 dinucleotide repeat motifs (88.68%), 18,363 trinucleotide repeat motifs (6.35%), 6,059 tetranucleotide repeat motifs (2.10%), 384 pentanucleotide repeat motifs (0.13%), and 444 (0.15%) hexanucleotide repeat motifs (Figure 3). For mononucleotide SSRs, the (A/T) n repeat was more prevalent and was about five times of the (C/G) n repeat, especial at the 0-10 bp length. The most abundant repeat in the dinucleotide SSRs was the (AG/CT) n repeat followed by the (AC/GT) n repeat. Both of these repeats accounted for 98.62% of the dinucleotides characterized (Additional file 3). For trinucleotide SSRs, the (AAC/GTT) n, (AAG/ CTT) n, and (CCG/CGG) n repeats were predominant, representing 27.24%, 19.28%, and 11.54%, respectively, of the trinucleotides identified (Additional file 4). Thirty-one tetranucleotide repeat motifs were recognized, and the most prevalent repeats were ACAT/ATGT (40.37%), ACGC/CGTG (15.23%), and ACTC/AGTG (12.02%) (Additional file 5). Together, pentanucleotide and hexanucleotide repeat motifs comprisedonly 0.29% of the total SSRs detected. The dominant pentanucleotide and hexanucleotide motifs were AGCTC/ AGCTG (10.42% of all of the pentanucleotide motifs) and ACACCC/ GGGTGT (43.47% of all of the hexanucleotide motifs), respectively (Additional files 6, 7, and 8).
Compound SSR analysis
In this study, 43,100 compound SSRs were identified, and these compound SSRs accounted for only 18.97% of all SSR sequences. Two types of compound SSRs were identified: one kind was without an interruption between the two motifs (C type; i.e., (AC)11taacactactcacacaaacacacacactctctcag (AC)10tcacact(CA)6) and another was with an interruption between the two motifs (C*type; i.e., (CT)16(TCT)5). In total, 40,464 C type (93.88%) and 2,636 C*type (6.12%) compound SSRs were detected. These results indicated the complexity of the broomcorn millet genome.
Validation identification of SSR markers and genetic diversity study of broomcorn millet in China
To identify the feasibility of using SSR sequences to study genetic diversity, 240 SSR primers were randomly choosed to synthesized and propagated through polymerase chain reaction to determine band polymorphisms in 40 broomcorn millet genotypes from diverse geographical locations. The result of PCR amplication showed that 103 SSR primer pairs generated a reproducible and distinct amplicon product on the specific size. Of these primer pairs, 66 (27.5%) were confirmed to amplify polymorphic bands from the 40 genotypes assessed (Table 2). Polymorphic variations in the SSRs were evaluated and listed in Table 3. The Na value (number of alleles) per locus changed from 2 to 5 with a mean of 2.67. The Ng value (number of amplified genotypes) ranged from 3 to 15 with an average of 5.21. The He (expected heterozygosity) and Ho (observed heterozygosity) varied from 0.05 to 0.77 (mean=0.45) and from 0 to 0.95 (mean=0.23), respectively. The mean I (Shannon Information index) value was 0.73 which varied from 0.11 to 1.52 per locus. These results indicated that SSR identification via next-generation sequencing is feasible and efficient. These SSRs can be used for future studies of broomcorn millet genetics.
| Primer pair ID | Ng | Na | I | Ho | He |
|---|---|---|---|---|---|
| ICSBM2 | 10 | 4 | 1.225 | 0.830 | 0.693 |
| ICSBM3 | 6 | 3 | 1.098 | 0.435 | 0.670 |
| ICSBM5 | 3 | 2 | 0.689 | 0.136 | 0.499 |
| ICSBM8 | 10 | 4 | 1.289 | 0.288 | 0.706 |
| ICSBM10 | 3 | 2 | 0.693 | 0.026 | 0.503 |
| ICSBM13 | 6 | 3 | 0.585 | 0.034 | 0.307 |
| ICSBM14 | 3 | 2 | 0.556 | 0.102 | 0.371 |
| ICSBM15 | 3 | 2 | 0.522 | 0.091 | 0.341 |
| ICSBM19 | 3 | 2 | 0.529 | 0.080 | 0.347 |
| ICSBM21 | 6 | 3 | 0.952 | 0.552 | 0.565 |
| ICSBM22 | 3 | 2 | 0.692 | 0.091 | 0.502 |
| ICSBM24 | 6 | 3 | 0.682 | 0.023 | 0.372 |
| ICSBM27 | 10 | 4 | 1.005 | 1.000 | 0.589 |
| ICSBM29 | 3 | 2 | 0.692 | 0.057 | 0.501 |
| ICSBM30 | 3 | 2 | 0.109 | 0.000 | 0.045 |
| ICSBM31 | 10 | 4 | 1.180 | 0.322 | 0.632 |
| ICSBM33 | 10 | 4 | 1.340 | 0.955 | 0.731 |
| ICSBM35 | 6 | 3 | 0.830 | 0.205 | 0.500 |
| ICSBM39 | 10 | 4 | 1.346 | 0.886 | 0.735 |
| ICSBM40 | 15 | 5 | 1.519 | 1.000 | 0.771 |
| ICSBM43 | 6 | 3 | 0.574 | 0.109 | 0.310 |
| ICSBM46 | 6 | 3 | 0.594 | 0.114 | 0.315 |
| ICSBM49 | 3 | 2 | 0.646 | 0.125 | 0.458 |
| ICSBM51 | 10 | 4 | 0.627 | 0.094 | 0.295 |
| ICSBM54 | 6 | 3 | 0.692 | 0.046 | 0.389 |
| ICSBM59 | 6 | 3 | 0.539 | 0.034 | 0.287 |
| ICSBM60 | 6 | 3 | 0.280 | 0.000 | 0.129 |
| ICSBM67 | 6 | 3 | 0.269 | 0.000 | 0.120 |
| ICSBM68 | 3 | 2 | 0.556 | 0.057 | 0.371 |
| ICSBM71 | 3 | 2 | 0.693 | 0.058 | 0.503 |
| ICSBM78 | 3 | 2 | 0.684 | 0.068 | 0.494 |
| ICSBM80 | 6 | 3 | 0.880 | 0.330 | 0.514 |
| ICSBM81 | 3 | 2 | 0.677 | 0.071 | 0.488 |
| ICSBM85 | 10 | 4 | 1.019 | 0.215 | 0.581 |
| ICSBM86 | 6 | 3 | 1.097 | 0.852 | 0.670 |
| ICSBM90 | 3 | 2 | 0.419 | 0.023 | 0.253 |
| ICSBM96 | 3 | 2 | 0.366 | 0.080 | 0.211 |
| ICSBM99 | 3 | 2 | 0.659 | 0.080 | 0.469 |
| ICSBM100 | 3 | 2 | 0.330 | 0.000 | 0.185 |
| ICSBM107 | 3 | 2 | 0.676 | 0.023 | 0.486 |
| ICSBM109 | 3 | 2 | 0.494 | 0.016 | 0.317 |
| ICSBM113 | 3 | 2 | 0.692 | 0.114 | 0.502 |
| ICSBM119 | 3 | 2 | 0.693 | 0.138 | 0.503 |
| ICSBM120 | 6 | 3 | 1.081 | 0.193 | 0.659 |
| ICSBM123 | 6 | 3 | 0.683 | 0.636 | 0.452 |
| ICSBM126 | 3 | 2 | 0.693 | 0.011 | 0.503 |
| ICSBM127 | 6 | 3 | 0.463 | 0.091 | 0.228 |
| ICSBM130 | 6 | 3 | 0.532 | 0.136 | 0.273 |
| ICSBM132 | 3 | 2 | 0.649 | 0.159 | 0.459 |
| ICSBM135 | 3 | 2 | 0.388 | 0.057 | 0.229 |
| ICSBM139 | 3 | 2 | 0.562 | 0.023 | 0.377 |
| ICSBM144 | 3 | 2 | 0.249 | 0.023 | 0.128 |
| ICSBM151 | 6 | 3 | 0.561 | 0.034 | 0.290 |
| ICSBM159 | 6 | 3 | 0.936 | 0.897 | 0.583 |
| ICSBM165 | 3 | 2 | 0.234 | 0.011 | 0.118 |
| ICSBM172 | 6 | 3 | 1.071 | 0.818 | 0.653 |
| ICSBM180 | 3 | 2 | 0.612 | 0.102 | 0.423 |
| ICSBM197 | 6 | 3 | 1.028 | 0.609 | 0.623 |
| ICSBM205 | 3 | 2 | 0.552 | 0.000 | 0.369 |
| ICSBM206 | 3 | 2 | 0.633 | 0.094 | 0.448 |
| ICSBM212 | 3 | 2 | 0.649 | 0.000 | 0.459 |
| ICSBM216 | 6 | 3 | 0.707 | 0.188 | 0.389 |
| ICSBM219 | 3 | 2 | 0.685 | 0.511 | 0.495 |
| ICSBM227 | 10 | 4 | 1.331 | 0.796 | 0.729 |
| ICSBM234 | 3 | 2 | 0.693 | 0.277 | 0.503 |
| ICSBM235 | 10 | 4 | 1.248 | 0.309 | 0.699 |
| ICSBM239 | 3 | 2 | 0.672 | 0.000 | 0.482 |
| 5.209 | 2.672 | 0.725 | 0.235 | 0.445 | |
| 2.766 | 0.786 | 0.302 | 0.300 | 0.173 |
Table 3: Polymorphic variationsin SSR loci following amplification of 40 geographically diverse Panicum miliaceum L. accessions. 1Ng: genotype No.; 2Na: observed number of alleles; 3I: Shannon information index; 4Ho: observed heterozygosity; 5He: expected heterozygosity.
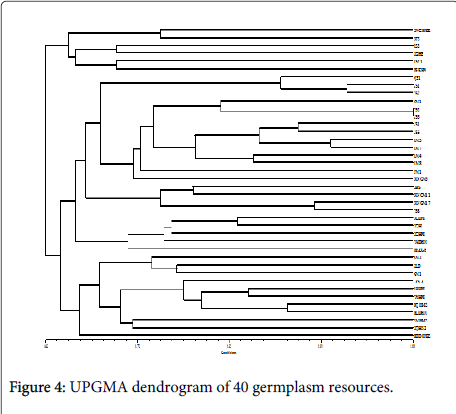
Cluster analysis was performed on the 40 broomcorn millet accessions using the UPGMA method according to Nei’s genetic distance theory. The dendrogram indicated when the genetic similarity value was 0.645, the 40 broomcorn millet accessions grouped into three different clusters (Figure 4). Cluster 1 had six accessions, including a series of Longshu varieties and their parents, which originated from Heilongjiang. Cluster 2 consisted of accessions mainly from Shanxi (7), Gansu (4), and Ningxia (5) as well as several accessions from Inner Mongolia (5). Cluster 3 had eleven accessions, including most (8) of the fourteen varieties collected from Inner Mongolia. Two accessions in Cluster 3 originated from Shanxi. This pattern of diversity was in accordance with the result reported by Hu et al.
Discussion
Broomcorn millet is a native vital crop for food safety in arid and semiarid areas
Broomcorn millet is an ancient domesticated crops, and its oldest historical reports date to 10,000-8,000 BC [32]. This crop was mention in nine poems of the ancient Chinese “Book of Poetry” (Shih Ching) and was regarded as most important grain in ancient [33]. Before rice and wheat became popular, Panicum was a staple food in countries of Eastern Asia, and then spread to the entire Eurasian continent [32,34-36]. Until today, Panicum remains an important food in these regions [37,38]. Due to its shorter growth cycle and resistance to salt, alkali, and drought stresses, broomcorn millet is usually grown as an exploratory cereal in new wastelands and deserts or as a remediation crop in the outburst of natural disasters. Previous researchers have identified large phenotypic variation in broomcorn millet germplasms [10], and using these rich genetic resources to improve broomcorn millet productivity is a promising field of study. However, genomic resources and data or high-polymorphism molecular markers of broomcorn millet is lacking for genetic analysis and this crop was regarded as a “genomic orphan”. Therefore, the development of userfriendly and highly polymorphic molecular markers is vital for the success of broomcorn millet breeding programs [39].
Development of microsatellite markers using 454 pyrosequencing
Microsatellites, is also called “short tandem repeats” or “simple sequence repeats”, distributed in Eukaryotic genomes uniformly, and were composed by tandem repeats of 1-6 nucleotides. There have abundant variation in repeats number of SSR between varieties or populations, as well as highly codominant inheritance, so SSR are DNA markers can be used frequently in many genomic researches [14]. Although the application of microsatellites is simple and robust, the identification and development of microsatellites is highly challenging [19]. There are two traditional approaches to SSR loci development. The first approach involves the de novo construction of a genomic library and microsatellite development, and the second approach involves testing microsatellite primers previously developed for related species. Microsatellite development methods based on the Sanger sequencing are often low throughput and typically only obtain a few hundred sequences due to the expensive of Sanger sequencing [15]. In addition, enrichment libraries generally constructed by several specific tandem repeats that selected randomly and no prior abundance information in the genome [40], this may be lead to bias of genome representation. 454 GS-FLX technology (next-generation sequencing) provides new opportunities for microsatellite isolation due to its high throughput, low cost of operation, and more thorough representation of the genome [15,41]. To date, several crops have developed high-throughput and novel genomic SSR markers via 454 GS-FLX sequencing [18-21,42]. In this study, massively parallel sequencing technology was adopted and hoped to discovery numerous SSR with high quality from genome of broomcorn millet quickly. A total of 1,087,428 high-quality broomcorn millet genomic unigenes were generated and the average length was 370 bp. The MISA analysis results showed that 223,894 SSRs were identified from 904,311 reads. Of the SSR-containing reads, mononucleotide, dinucleotide, and trinucleotide repeat motifs are dominated in the broomcorn millet genomic sequences. This result is Similar to the findings in other crops [21,43]. The (AG/CT) n repeat was accounting fosr 45.8% of total identified SSRs, and was the predominant repeat motif in the whole genome of broomcorn millet. The (AG/CT) n repeat was followed in abundance by (AC/GT) n, (A/T) n, and (AAC/GTT) n. In this study, (AAT/ATT)n, (ATC/ATG)n, and (ACT/AGT)n were rarely detected. The isolation and identification of unwanted repeat motifs, such as (AAT/ATT)n, (ATC/ATG)n, and (ACT/AGT)n, which are present in low proportions, should enhance the number of successful primers designed.
SSRs can be effective molecular markers for the genomic analysis of broomcorn millet
SSR markers can link the genotypic and phenotypic variation, this may be helpful for breeders to expedite the development of improved cultivars [44]. In addition, SSR markers are fit for the genetic linkage map construction, genetic diversity analysis, QTL mapping, gene cloning, and marker-assisted selection due to their widely distribution in eukaryotic genomes [45]. Next generation sequencing technology can be used for the high-throughput identification of SSR loci in crop species, especially in “orphan” crop species that lack available genetic and genomic resources [46]. In this paper, 454 pyrosequencing was chose to develop codominant and polymorphic genetic SSR markers in broomcorn millet. This method was highly effective as 103 of the 240 (42.9%) randomly designed SSR primers successfully amplified stable and clear bands after PCR reaction. Then the 103 markers were selected for genetic diversity and group delineation of 40 broomcorn millet accessions which originate from 5 different ecotypes, we validated that 66 markers were highly polymorphic in test accessions.
The polymorphic markers enhanced our cognization about the genetic diversity level of broomcorn millet accessions. For broomcorn millet, isozymes and protein electrophoresis for intra-species grouping or classification have not been successful [1,47]. Genomic SSR markers in broomcorn millet were also identified more efficiently than the transferred SSR markers which selected from other crops. Hu et al. reported that only 46 of the 983 SSR primer pairs (4.6%) tested could generate clear and reproducible polymorphic fragments [12]. This numerous genomic SSR markers developed in the study will facilitate the evaluation of genetic structure and the construction of highresolution maps in broomcorn millet.
Conclusion
This study provides a broad discovery and characterization of microsatellites loci in the broomcorn millet genome using 454 GS FLX Titanium sequencing technology. Moreover, massive SSR-enriched sequence data were first generated, facilitating the discovery and utilization of genomic SSR markers, further to accelerate the genomic and genetic research of broomcorn millet.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant No.31301386), the National Millet Crops Research and Development System (NMCRDS), China Agriculture Research System (CARS-07-12[1].5-A1) and the Agricultural Science and Technology Innovation Program (ASTIP) in CAAS. We are grateful to Dr. Dahai Wang and Liping Sun (Beijing Autolab Biotechnology Co., Ltd) for special contribution to this work.
References
- M'Ribu HK, Hilu KW (1994) Detection of interspecific and intraspecific variation in panicum millets through random amplified polymorphic DNA. Theor Appl Genet 88: 412-416.
- Hunt HV, Badakshi F, Romanova O, Howe CJ, Jones MK, et al. (2014) Reticulate evolution in panicum (Poaceae): The origin of tetraploid broomcorn millet, P Miliaceum. J Exp Bot 65: 3165-3175.
- Crawford GW, Zhang J, Liu KB, Wu N, Li Y, et al. (2009) Agricultural origins in north China pushed back to the Pleistocene-Holocene boundary. Proc Natl Acad Sci USA 106: 7271-7272.
- Dong Y, Zheng D (2006) Crops and their wild relatives in China. China Agriculture Press, Beijing.
- Wang L, Wang XY, Wen QF, Cao LP (2007) Identification on protein and fat content for Chinese proso millet germplasm resources. Journal of Plant Genetic Resources 8: 165-169.
- Liu MX, Zhang ZW, Wu B, Lu P (2012) Evaluation of mixed salt-tolerance at germination stage and seedling stage and the related physiological characteristics of panicum miliaceum L. Scientia Agricultura Sinica 45: 3733-3743.
- Baltensperger DD (2002) Progress with proso, pearl and other milllets. In: Janick J, Whipkey A (eds.). Trends in new crops and new uses. ASHS Press, Alexandria, pp: 100-103.
- Wang XY, Wang L, Wen QF (2004) Identification of broomcorn millet varieties resource for resistance to smut in China. Journal Shihezi University 7: 43-45.
- Lágler R, Gyulai G, Humphreys M, Szabó Z, Horváth L, et al. (2005) Morphological and molecular analysis of common millet (P. Miliaceum) cultivars compared to a DNA sample from the 15th century (Hungary). Euphytica 146: 77-85.
- Wang L, Wang XY, Wen QF (2005) Research and utilization of proso millet germplasm resource in China. Journal of Plant Genetic Resources 6: 471-474.
- Hunt HV, Campana MG, Lawes MC, Park YJ, Bower MA, et al. (2011) Genetic diversity and phylogeography of broomcorn millet (panicum miliaceum L.) across Eurasia. Mol Ecol 20: 4756-4771.
- Hu X, Wang J, Lu P, Zhang H (2009) Assessment of genetic diversity in broomcorn millet (panicum miliaceum L.) using SSR markers. J Genet Genomics 36: 491-500.
- Cho Y, Chung J, Lee G, Ma K, Dixit A, et al. (2010) Development and characterization of twenty-five new polymorphic microsatellite markers in proso millet (panicum miliaceum L.). Genes Genomics 32: 267-273.
- Sunnucks P (2000) Efficient genetic markers for population biology. Trends Ecol Evol 15: 199-203.
- Malausa T, Gilles A, Meglécz E, Blanquart H, Duthoy S, et al. (2011) High-throughput microsatellite isolation through 454 GS-FLX Titanium pyrosequencing of enriched DNA libraries. Mol Ecol Resour 11: 638-644.
- Lam HM, Xu X, Liu X, Chen W, Yang G, et al. (2010) Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet 42: 1053-1059.
- Huang X, Wei X, Sang T, Zhao Q, Feng Q, et al. (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42: 961-967.
- Tangphatsornruang S, Somta P, Uthaipaisanwong P, Chanprasert J, Sangsrakru D, et al. (2009) Characterization of microsatellites and gene contents from genome shotgun sequences of mungbean (Vignaradiata (L.) Wilczek). BMC Plant Biol 9: 137.
- Csencsics D, Brodbeck S, Holderegger R (2010) Cost-effective, species-specific microsatellite development for the endangered dwarf bulrush (Typha minima) using next-generation sequencing technology. J Hered 101: 789-793.
- Yang T, Bao SY, Ford R, Jia TJ, Guan JP, et al. (2012) High-throughput novel microsatellite marker of faba bean via next generation sequencing. BMC Genomics 13: 602.
- Yang T, Jiang J, Burlyaeva M, Hu J, Coyne CJ, et al. (2014) Large-scale microsatellite development in grasspea (Lathyrussativus L.), an orphan legume of the arid areas. BMC Plant Biol 14: 65.
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349.
- Kandpal RP, Kandpal G, Weissman SM (1994) Construction of libraries enriched for sequence repeats and jumping clones, and hybridization selection for region-specific markers. Proc Natl Acad Sci USA 91: 88-92.
- Glenn TC, Schable NA (2005) Isolating microsatellite DNA loci. Methods Enzymol 395: 202-222.
- Rice P, Longden I, Bleasby A (2000) EMBOSS: the European molecular biology open software suite. Trends Genet 16: 276-277.
- Rozen S, Skaletsky H (2000) Primer3 on the www for general users and for biologist programmers. Methods Molbiol 132: 365-386.
- Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106: 411-422.
- R Core Team (2012) R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
- Yeh FC, Yang RC, Boyle T (1999) POPGENE software package version 1.31 for population genetic analysis. University of Alberta.
- Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70: 3321-3323.
- Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76: 5269-5273.
- Crawford G (2005) East Asian plant domestication. Archaeology of Asia. In: Stark MT (eds.), Blackwell Publishing House, Malden, pp: 77-95.
- Keng H (1974) Economic plants of ancient north China as mentioned in Shih Ching (Book of Poetry). Econ Bot 28: 391-410.
- Bellwood P (2005) First Farmers: the origins of agricultural societies. Blackwell Publishing House, Malden, pp: 87-109.
- Zohary D, Hopf M (2000) Domestication of plants in the old word: the origin and spread of cultivated plants in West Asia, Europe and the Nile valley. Oxford University Press, Oxford, UK.
- Fuller DQ (2006) Agricultural origins and frontiers in South Asia: A working synthesis. J World Prehist 20: 1-86.
- You XL (1993) The question for origin and spread in both foxtail millet and common millet. Agric Hist (China) 12: 1-13.
- Li Y, Wu S (1996) Traditional maintenance and multiplication of foxtail millet (Setaria italica (L.) P. Beauv.) landraces in China. Euphytica 87: 33-38.
- Collard BC, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc Lond B Biol Sci 363: 557-572.
- Castoe TA, Poole AW, Gu W, Jason de Koning AP, Daza JM, et al. (2010) Rapid identification of thousands of copperhead snake (Agkistrodoncontortrix) microsatellite loci from modest amounts of 454 shotgun genome sequence. Mol Ecol Resour 10: 341-347.
- Santana Q, Coetzee M, Steenkamp E, Mlonyeni O, Hammond G, et al. (2009) Microsatellite discovery by deep sequencing of enriched genomic libraries. BioTechniques 46: 217-223.
- Zhu H, Senalik D, McCown BH, Zeldin EL, Speers J, et al. (2012) Mining and validation of pyrosequenced simple sequence repeats (SSRs) from American cranberry (VacciniummacrocarponAit.). Theor Appl Genet 124: 87-96.
- Shi J, Huang S, Fu D, Yu J, Wang X, et al. (2013) Evolutionary dynamics of microsatellite distribution in plants: insight from the comparison of sequenced brassica, Arabidopsis and other angiosperm species. PLOS ONE 8: e59988.
- Gupta PK, Varshney RK (2000) The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 113: 163-185.
- Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet 30: 194-200.
- Martinez S, Correal E, Real D, Ortuño A, Del Río J (2010) Bituminariabituminosa: a source of furanocoumarins of pharmaceutical interest. In: Awaad AS, Govil JN, Singh VK, (eds.). Recent progress in medicinal plants (RPMP). Drug plants, pp: 307-322.
- Warwick SI (1987) Isozyme variation in proso millet. J Hered 78: 210-212.
Relevant Topics
- Agricultural science
- Agronomy
- Climate impact on crops
- Crop Productivity
- Crop Sciences
- Crop Technology
- Field Crops Research
- Hybrid Seed Technology
- Irrigation Technology
- Organic Cover Crops
- Organic Crops
- Pest Management
- Plant Genetics
- Plant Breeding
- Plant Nutrition
- Seed Production
- Seed Science and Technology
- Soil Fertility
- Weed Control
Recommended Journals
Article Tools
Article Usage
- Total views: 3916
- [From(publication date):
August-2017 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 3068
- PDF downloads : 848