Development of Phage Synthesizing System by the Minimizing Approach
Received: 16-Oct-2018 / Accepted Date: 18-Oct-2018 / Published Date: 22-Oct-2018 DOI: 10.4172/2332-0877.1000383
Keywords: Bacteriophage; Virion; Genome
Introduction
The synthesis of life has been the vision of mankind, and some researchers are attempting it [1]. I consider the creation of life to the almost same as understanding life. I also consider the concept of life is quasi-equal to viruses. I tried to construct a phage synthesizing system based on PURE SYSTEM (Shimizu et al 2001) [2]. Further the reason I selected the minimizing approach is that understanding of “life” to be the division of the real chaotic world into smaller ones.
The MS2 bacteriophage used this study has a single stranded RNA genome that encodes 4 genes : A (maturation), coat, replicase, and lysis. The diameter of this phage is 26nm, and its protein shell is 2 nm wide. The A protein is required for attachment to pili (infection); each virion has one copy of the A gene, and this protein is translated at a low level. The replicase protein is required for the replication of the RNA genome. The lysis protein causes cell lysis. The 180 copies of coat protein form an icosahedral particle. The A-RNA genome complex can infect bacteria that have pilli (Shiba and Miyake, 1975) [3]. Hence, I considered this complex to entity of the phage.
Materials and Methods
MS2 RNA isolation MS2 phages were grown on Escherichia coli strain XL-Blue, and phage RNA was extracted by phenol/chloroform treatment.
Translation of MS2 RNA the MS2 RNA genome was translated using PURE SYSTEM and the S30 extract system for linear templates (PROMEGA). Translation mixtures in a volume of 30μl were incubated at 37°C. Protein synthesis was monitored by SDS-PAGE, in which 15% acrylamide gels were used.
Phage assay F+ E. coli XL1-Blue was grown in LB medium under aerobic conditions until a biomass concentration OD500 (0.5) was achieved. Next, 200 μl of this suspension was mixed with 1μl of the translation mixture and 3 ml of melted top agar (47°C) and poured onto plates containing hardened bottom agar. The numbers of plaques were counted after 8-12 h of incubation at 37°C.
Identification of the plaque - forming factor
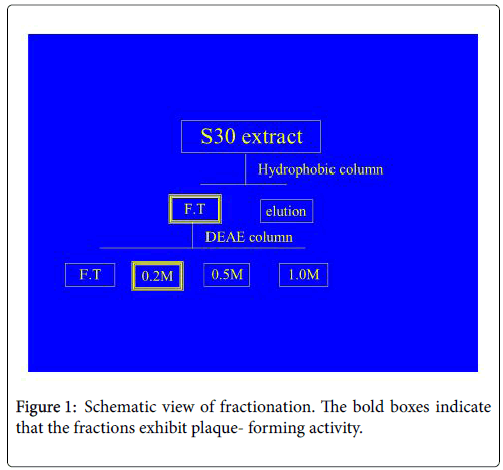
The procedure used is similar to that used by Momose et al. (1988) [4] (Figure 1). The buffer used for the fractionation of the plaque – forming factor contains 1 M HEPES-KOH (pH7.6) and 10% (v/v) glycerole (buffer G), this buffer was prepared both with and without KCl. The S30 extract system was purchased from PROMEGA. The S30 extract was loaded onto a hydrophobic column (Butyl-650M) equilibrated with Tris buffer (pH 7.6). And the material that was not absorbed to the column was onto DEAE anion exchange column equilibrated with buffer G containing 0.05M KCl. The material absorbed on the column was eluted stepwise with buffer G containing 0.2 M, 0.5 M, and 1.0 M KCl. The plaque – forming units were recovered in the fraction eluted with 0.2 M KCl.
Results
When the genomic RNAF of MS2 is treated with PURE SYSTEM and the S30 extract system are produced phage proteins; the most abundant of these is the coat protein.
The β- subunits of the RNA replicase and maturation protein were also synthesized. The translation reaction mixture was assayed using F + E. coli XL-1 Blue. The S30 translation mixture produced viable plaque – forming units on the bacterial low Vladimir L. Katanaev, et al (1996) [5]. PURE SYSTEM was not or very little. I hypothesized that S30 contains some factors related to the formation of the infectious units.
To test this hypothesis, I added 2μl of ultra filtered S30 to 30 μl of the PURE SYSTEM mixture, and I used this solution to the MS2 genome. Merely 1μl of this translation mixture could infect F+ E. coli .
I considered the hypothesis to be correct. In order to identify the unique factors in S30, I used a hydrophobic column to fractionate the S30 extract, I corrected the flow-through and elution fractions. Infectious units were synthesized when 30 μl of the PURESYSTEM mixture was mixed with 2μl of the flow-through fraction. Next, I used a DEAE anion-exchange column for further investigation. 0.2 M KCl fraction had the ability to form plaque forming units. I measured the absorbance of this fraction, and the absorbance peak was at around 260 nm. This finding indicates that the factors present in this fraction are nucleic acids.
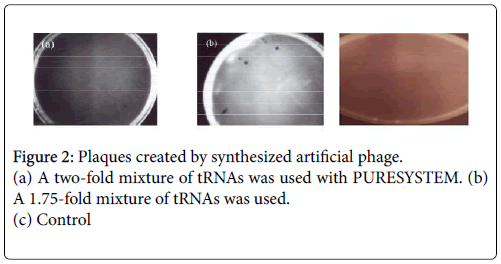
I considered whether or not these factors could be tRNAs. Elder and Smith mentioned that tRNAs are present in the virion particle of the avian tumor virus (1973) [6]. I added tRNAs to the PURE SYSTEM mixture. Many plaques appeared on the bacterial lawns (Figure 2), indicating that the factors responsible for infectious unit formation are tRNAs.
Discussion
I successfully established a minimum phage synthesizing system. The system is under non-equilibrium state. I also precisely identified the factors related to the synthesis of plaque-forming units. In this study, I showed that it is possible to create life from very simple organic materials. This system may be applicable to other pathogenic viruses, and it could be used to synthesize novel drugs.
Acknowledgments
I thank Drs Y. Shimizu, H. Taguchi, and T. Ueda for providing important suggestions. I also thank Dr Jan van Duin for recommending many articles and giving valuable advise.
References
- Szostak JW, Bartel DP, Luisi PL (2001) Synthesizing life. Nature 409: 387-390.
- Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, et al. (2001) Cell-free translation reconstituted with purified components. Nat Biotechnol 19: 751-755.
- Shiba T, Miyake T (1975) New type of infectious complex of E. coli RNA phage. Nature 254: 157-158.
- Momose F, Handa H, Nagata K (1996) Influenza Virology Current Topics. Biochimie 78: 1103-1108.
- Katanaev VL, Spirin AS, Reuss M, Siemann M (1996) Formation of bacteriophage MS2 infectious units in a cell-free translation system. FEBS Lett 397: 143-148.
- Elder KT, Smith AE (1973) Current Topics in Microbiology and Immumology. Nature 247, 435-438.
Citation: Ogimura H (2018) Development of phage synthesizing system by the minimizing approach. J Infect Dis Ther 6: 383. DOI: 10.4172/2332-0877.1000383
Copyright: © 2018 Ogimura H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 2725
- [From(publication date): 0-2018 - Nov 21, 2024]
- Breakdown by view type
- HTML page views: 2132
- PDF downloads: 593