Research Article Open Access
Development of Microbial Consortia for the Effective Treatment of Complex Wastewater
| Ghyandeep L. Gaikwad1*, Satish R. Wate1, Dilip S. Ramteke1 and Kunal Roychoudhury2 | |
| 1 CSIR-National Environmental Engineering Research Institute, Nehru Marg, Nagpur, India | |
| 2 S. K. Porwal College, Kamptee, Dist. Nagpur, India | |
| Corresponding Author : | Ghyandeep L. Gaikwad Environmental Impact and Risk Assessment Division CSIR- National Environmental Engineering Research Institute [CSIR- NEERI] Nehru Marg, Nagpur, India Tel: 919423107133, +917122249896 Fax: +917122249896 E-mail: swapn_gaik@rediffmail.com |
| Received January 03, 2014; Accepted May 14, 2014; Published May 18, 2014 | |
| Citation: Gaikwad GL, Wate SR, Ramteke DS, Roychoudhury K (2014) Development of Microbial Consortia for the Effective Treatment of Complex Wastewater. J Bioremed Biodeg 5:227. doi:10.4172/2155-6199.1000227 | |
| Copyright: © 2014 Gaikwad, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
The complex wastewater was characterized for physicochemical parameter. Five microbial species were isolated from different potential sources and their degradation potential was evaluated individually and in the form of consortia. Accordingly, lab studies were carried out in biological treatment system using aerobic microbial consortia along with individual microbial species in separate reactors. Temperature and pH of reaction tank was continuously monitored during study period. Other vital physicochemical parameters viz. COD, BOD, TSS and TDS were evaluated during 72 hours of study. The microbial consortia comprise of Pseudomonas spp., Actinomycetes spp., Bacillus spp., Streptomyces spp. and Staphylococcus spp. was able to trim down the physicochemical parameters of complex wastewater. The consortia demonstrated high COD and BOD reduction up to 90.17% and 94.02% respectively, compared to individual microbial species ranging from 42.11-59.76% for COD and 58.55-77.31% in case of BOD.
One of the challenging issues facing mankind at this juncture is that of pollution of freshwater by various anthropogenic activities. Industries are consuming large amount of water and release large amount of effluent loaded with many pollutants [4]. In India more than 55% of the industries do not have any type of effluent treatment plant and around 20% have partial treatment facilities. All of these industries drain off black liquor without chemical recovery or treatment of effluent due to economic reasons [5]. In addition, microbial contamination from inadequate sanitation facilities, improper wastewater disposal, and animal wastes leads to the greatest effect on human health [6,7]. Thus, the current scenario is such that the higher concentration of contaminants even in the treated effluents, reach the surface and ground water systems thereby affecting vegetation, humans and animals. [5].
The industrial, municipal and agricultural wastes, which are legally or illegally discharged into the environment, are responsible for environmental pollution [8-10]. The major pollutants are present in the form of black liquor [4,11]. These effluents are dark brown in colour and associated with high BOD, COD, total solids and organic carbon [12]. The value of BOD, TSS and TDS varies with respect to industries. The industry specific BOD values are, for tanning industry (959.66 mg/lit), Textile industry (3900 mg/lit), Dairy Industry (8239 mg/lit), Cheese industry (mg/lit), Tannery (1100-2500 mg/lit), Yogurt and Buttermilk (1000 mg/lit), Distillery wastewater (36,000-204,000 mg/lit) [13-20].
Naturally occurring microorganisms are the workhorses of wastewater treatment consisting of bacteria, fungi, protozoa, rotifers, and other microbes. These organisms thrive on many of the complex compounds contained in wastewater. Small size, high surface area-to-volume ratio, and large contact interfaces with their surrounding environment are some of the ideal features of microorganisms as bio-indicators of chemical pollutant stressors [21].
The treatment of wastewater is an immense challenge now days. Though conventional wastewater treatment plants are running, it is very difficult for them to cope up the norms devised by regulatory authorities. Accordingly, the dire need has generated to develop the new technology for effluent treatment or to make the appropriate modification in the existing treatment process. The present study thus intends to perform by keeping the view that the conventional activated sludge process be boosted by introducing new kind of microbial flora along with indigenous one.
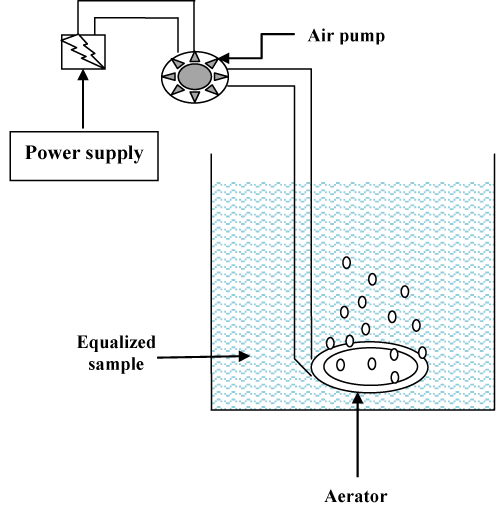
The vital parameters with respect to wastewater treatment viz. Temperature, pH, Total Suspended Solid (TSS), Total Dissolved Solid (TDS), Biochemical Oxygen Demand (BOD) and Chemical Oxygen Demand (COD) were evaluated during the time of study. Treatability study was also performed using consortia and the same parameters were evaluated. The results are shown in table 4.
The six crucial parameters were studied intermittently up to 72 hours when a microbial consortium was used in biological treatment system. The data obtained from the experimental setup illustrate that temperature of the system varies by 1°C in 72 hours span of the experiment. The 1°C fluctuation in temperature may not cause any deleterious effect on the life of microbial population in reactor and thus consortia contribute the degradation process of recalcitrant present in effluent. The pH of the system lies almost neutral during entire course of study which facilitates the growth of microbial consortia in wastewater.
The high Total suspended solids (TSS) can cause an increase in surface water temperature, because the suspended particles absorb heat from sunlight. This can cause dissolved oxygen levels to fall [23] and impart adverse effect on microbial life. It is known that warmer water holds less oxygen than cooler. The results also illustrates that total suspended solids was comparatively high at the zero hour however microbial consortium was able to reduce it by 79.76% within 72 hours in reactor. The value of TSS obtained after 72 hours came to permissible limit.
The total dissolved solid is closely links the bulk conductivity to microbial degradation of hydrocarbon [24]. The principal ions contributing to dissolved solids are carbonate, bicarbonate, chloride, sulphate, nitrate, silicates, sodium, potassium, calcium and magnesium. TDS value is observed to be reduced by 74.36% by microbial consortia after 72 hours. TDS value gradually decreases within time course of study by microbial consortia within permissible limit.
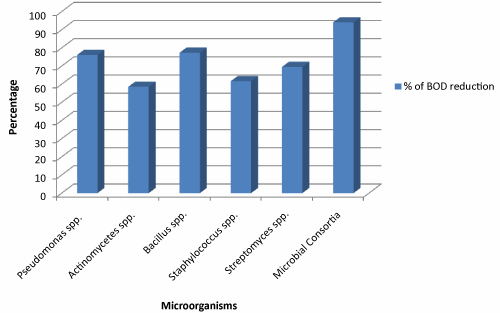
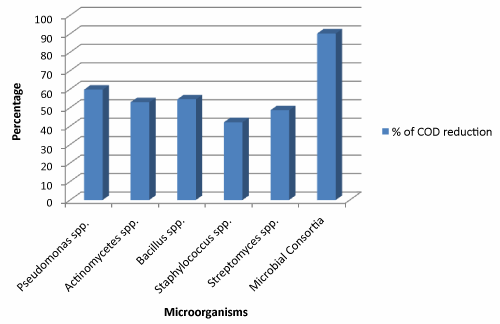
The microbial consortia comprises of Pseudomonas spp., Actinomycetes spp., Bacillus spp., Staphylococcus spp. and Streptomyces spp. showing optimistic effect in the reduction of vital content of wastewater viz. BOD and COD values. The Biochemical oxygen demand (BOD) is a measure of the oxygen used by microorganisms to decompose the waste. BOD was high at the zero hour of experiment but could be reduced below the permissible limit (100 mg/lit) as the time course of study progress. The maximum percentage removal of BOD in 72 hours by consortia was found to be 94.02% (Figure 2). Consortial strategy has been adopted by some workers for industry specific treatment. The BOD reduction of domestic wastewater by sedimentation, aeration, activated sludge and sand filter was reported as 97.66% using consortial system [25]. The value of Chemical Oxygen Demand (COD) at the onset of study was comparatively high. The microbial consortium was certainly able to reduce the COD value up to manifold. Figure 3 illustrate the percentage of COD reduction between microbial consortia and individual microbial species. The figure apparently suggests that microbial consortia able to reduce the COD by 90.17% compared to individual microbial species in the reaction tank. The other workers reported the reduction of COD in domestic sewage by 92.17% [25]. The percentage of COD reduction by individual microbial species reaches up to 59.76%, 52.95%, 54.53%, 42.11% and 48.66% by Pseudomonas spp., Actinomycete spp., Bacillus spp., Staphylococcus spp. and Streptomyces spp. respectively.
References
- Rangachari R (2005) Bhakra-Nangal Project, India: Regional and National Impact, New Delhi: Indian Water Resources Society.
- Biswas AK (2007) Water as a human right in the MENA Region: challenges and opportunities. International Journal of Water Resources Development 23: 209-225.
- Sinha, DK,Shrivastava AK (1995) Physico-chemical characteristics of river Sai at Raibareli. Indian Journal Environment Health 37: 205-210.
- Yelda, S, Mitra A, Bandyopadhyay M (2002) Purification of Pulp and Paper Mill Effluent Using EichorniaCrassipes. Environ Technol 23: 453-465.
- FAO (1999) The State of the Word’s forest. ISBN 92-5-104193-8. FAO Documentation Group Cataloguing in Publishing Data FAO, Rome,Italy.
- World Water Assessment Programme (2009) The United Nations World Water Development Report 3: Water in a Changing World, Paris: UNESCO; London: Earthscan.
- Corcoran E., Nellemann C,Baker E,Bos R,al. (2010) Sick Water? The Central Role of Wastewater Management in Sustainable Development: A Rapid Response Assessment, United Nations Environment Programme. ISBN: 978-82-7701 075-5. Printed by BirkelandTrykkeri AS, Norway.
- Bakare A, Lateef A, Amuda OS (2003) The aquatic toxicity in characterization of chemical and microbiological constituents of water samples from oba river, odo-oba, Nigeria. Asian J Microbial Biotech EnvSci 5: 11-17.
- Shashirekha V, Pandi M, Mahadeswara S (2005) Bioremediation of tannery effluents and chromium containing wastes using cyanobacterial species. J AmerLeathChem Ass 100: 419-426.
- Shashirekha V, Sridharan MR, Swamy M (2008) Biosorption of trivalent chromium by free and immobilized blue green algae: kinetics and equilibrium studies. J Environ Sci Health A Tox Hazard Subst Environ Eng 43: 390-401.
- Ingle ST (2000) Pollution Potential of Pulp and Paper Mill Effluent. Bull EnviSci 18:21- 24.
- Kirk TK, Jeffries TW, Leatham GF (1983) Biotechnology: Application and implications for the Pulp and Paper Industry. Tappi J 66: 45-51.
- Ayoub GM, Hamzeh A, Semerijan L (2011) Post treatment of tannery wastewater using lime/bittern coagulation and activated carbon adsorption. Desalination 273: 359-365.
- Ahn DH, Chang WS, Yoon TI (1999) dyestuff wastewater treatment using chemical oxidation, physical adsorption, and fixed bed biofilm process. Process Biochemistry5: 429-439.
- Arbeli A, Brenner Z, Abeliovich A (2006) Treatment of high-strength dairy wastewater in an anaerobic deep reservoir: Analysis of the methanogenicfermentaionin a moving bed biofilm reactor. Water sci. Technol 45: 321-328.
- Monroy OH, Guoyot JP, Vazquez JC, Derramadero FM (1995) Anaerobic-Aerobic treatment of cheese wastewater with national technology. Water Sci. Technol 32:149-156.
- RaghavaRao J, Chandrababu NK, Muralidharan C, Unni Nair B, Rao PG, et al. (2003) Recouping the wastewater: a way forward for cleaner leather processing. Journal of Cleaner Production 11:591-599.
- Koyuncu I, Topacik D, Turan M, Celik M, Sarikaya MS (2001) Influence of filtration conditions on the performance of nanofiltration and reverse osmosis membranes in dairy wastewater treatment. Water Sci. Technol 1:117-124.
- Piya-areetham P, Shenchunthichai K, Hunsam M (2006) Application of electrooxidation process for treating concentrated wastewater from distillery industry with a voluminous electrode. Water Research 40:2857-2864.
- Al-a'ama MS, Nakhila GF (1995) Wastewater Reuse in Jubail, Saudi Arabia. Water Research. 29: 1579-1584.
- Ramakrishnan B (2012) Microbial community tracking in bioremediation. J BioremedBiodeg 3:10.
- Rice EW, Bair RB, Eaton AD, Clesceri LS (2012) Standard methods, for the examination of water and wastewater, (22ndedn), ISBN 978-087553-013-0. ISSN 55-1979. Published jointly by American Public Health Association, American Water works Association and Water Environment Federation.
- Mitchell MK, Stapp WB (1992) Field Manual for Water Quality Monitoring, an environmental education program for schools. GREEN: Ann Arbor, MI.
- Eliot A. Atekwanaa, Estella A. Atekwanaa, Rebecca S. Roweb, D. Dale WerkemaJr.c, Franklyn D. Legall (2004) The relationship of total dissolved solids measurements to bulk electrical conductivity in an aquifer contaminated with hydrocarbon. Journal of Applied Geophysics, 56: 281-294.
- Al-Jlil S (2009) COD and BOD reduction of waste water using activated sludge sand filters and activated carbon in Saudi Arabia. Biotechnol 8: 473-477.
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 | Table 4 |
Figures at a glance
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 18077
- [From(publication date):
July-2014 - Nov 21, 2024] - Breakdown by view type
- HTML page views : 13089
- PDF downloads : 4988
