Development of a Saliva-based Lateral Flow Assay for SARS-CoV-2 with the potential to quantify viral load
Received: 01-Jun-2023 / Manuscript No. jabt-23-103065 / Editor assigned: 03-Jun-2023 / PreQC No. jabt-23-103065 / Reviewed: 16-Jun-2023 / QC No. jabt-23-103065 / Revised: 19-Jun-2023 / Manuscript No. jabt-23-103065 / Accepted Date: 25-Jun-2023 / Published Date: 26-Jun-2023 QI No. / jabt-23-103065
Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is one of the deadliest virus in the last 50 years, with the USA having the highest reported deaths and cases at over 1.1 million and over 100 million, respectively, as of June 1, 2023. Identifying infected people remains the primary method of stopping the spread of the virus, either using real-time, quantitative polymerase chain reaction instruments or at-home lateral flow assay (LFA) antigen tests. Herein we describe a simple four step at-home SARS-CoV-2 LFA test that provides three advantages over current LFAs. The test employs 1) saliva sampling, 2) three antibodies to bind the virus to the LFA Test Line, and 3) a smartphone to quantify the reflectance of the Test Line in terms of Ct values. The use of saliva samples eliminates the pain and fear of nasopharyngeal sampling, especially for children. The use of three antibodies yielded 100% correct sensitivity, specificity, predicted positive and predicted negative for samples with Ct values of 29 and below. The use of a smartphone to measure reflectance allowed calculating the Ct values for 16 samples with an average error and standard deviation of 0.58±0.43 for samples with Ct values below 26. The smartphone also adds the capability of sharing the results to track and slow the spread of the virus. The combination lateral flow assay and smartphone quantitation is well suited for identifying, quantifying, and slowing the spread of future viruses.
Keywords
SARS-CoV-2; Viral load; Saliva sampling
Introduction
In just over three years since the initial detection of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the USA, more than 1.1 million people have died and over 100 million cases have been reported, far more than any other country in the world [1]. A major component to stopping the spread of the disease was the development of real-time, quantitative polymerase chain reaction methods (PCR) used to identify infected people. PCR employs nucleic acid primers to double the amount of SARS-CoV-2 ribonucleic acid in an oral fluid or nasopharyngeal sample through each temperature cycle. The cycles are repeated until a fluorescent dye attached to the primers increases sufficiently to be detected, otherwise known as the cycles-to-threshold of detection (Ct). The Ct value is an indication of the concentration of the virus in the sample. Specific primers used to replicate the SARSCoV- 2 genome, identified in January 2020 [2], were incorporated into existing PCRs, and widespread testing began in March 2020 [3]. Tests sites were set up initially at hospitals, and later at many drive-through venues. By July 2020, the Center for Disease Control (CDC) established a national tracking database for all PCR test results [4]. A significant amount of testing was driven by employers who wanted healthy, noncontagious workers. Unfortunately, PCR has several limitations, such as the use of expensive primers, and the need for highly trained technicians and well-equipped laboratories. More importantly, the test takes 2-6 hours to perform and 2-5 days to provide results, due to limited PCR instrument availability and sample transport to labs. During the delay contagious people spread the virus to others [5]. In order to overcome these limitations, we and others developed at-home tests. Such tests would eliminate drive-through lines and the wait for results, and they cost much less than PCR tests. Our test, like most, employed antibodies to bind the virus at the Test Line of a lateral flow assay (LFA) [6]. However, there are two important differences. Saliva was used as the sample medium, and three antibodies were used to bind to the virus. The former proved useful in the testing of children, and the latter provided greater sensitivity compared to other LFA athome tests. Here we describe the development of the LFA, Food and Drug Administration (FDA) test results required for emergency use authorization, and the use of a smartphone to quantify the Test Line in terms of Ct values.
Materials and Methods
Materials
All reagents, such as HPLC water and pH 7 buffers, were obtained from Sigma-Aldrich (Allentown, PA), while COVID-19 antibodies were obtained from Meridian Bioscience (Cincinnati, OH). Heat inactivated SARS-Related Coronavirus 2, Isolate USA-WA1/2020, was obtained from BEI Resources (NR-52286, Lot: 70037779, Bethesda, MD). De-identified pooled saliva samples, with and without the SARS-CoV-2 virus for product development were obtained from Lee Biosolutions (Maryland Heights, MO). De-identified pooled saliva samples with the SARS-CoV-2 virus for quantitation were obtained from Boca Biolistics (Pompano Beach, FL). Viruses, bacteria, common drugs, and human blood were purchased from BEI Resources, American Type Culture Collection (ATCC, Manassas, Virginia), Hardy Diagnostics (Santa Maria, California), ZeptoMatrix (Buffalo, New York), and the University of Rhode Island (Kingston, RI) as indicated in Table 1. Microbial interferents, listed in Table 2, were obtained from Lee Biosolutions. All pathogenic samples were received in 2 mL plastic vials, and all sample preparations were performed in a Biosafety Level 2 cabinet following standard safety procedures, using non-cotton swabs, 1 mL plastic centrifuge tubes and a manual pipetter for sample and buffer transfer, all from VWR (Bridgeport, NJ ). The COVID-19 Saliva Antigen At-Home Test consisted of a polyester flock swab (Guilford, ME), a 2 mL plastic vial containing 0.5 mL of buffer, a 1 mL disposable plastic pipette (Franklin, MA), and an RTA designed lateral flow assay cassette (Figure 1A). The LFA cassettes were of standard construction (nanoComposix, San Diego, CA), consisting of 1) a sample pad, 2) a conjugate pad containing probes, 3) a Test Line functionalized with mouse monoclonal SARS-CoV-2 nucleocapsid and spike protein antibodies as capture antibodies, 4) a Control Line functionalized with goat anti-mouse IgG antibodies, and 5) a wicking pad, all on 6) a nitrocellulose support, and 7) enclosed in a plastic cassette containing a sample addition port and a viewing section. The conjugate probes consisted of synthesized gold nanoparticles [7], coated with a dye, as a reporter molecule, and were also functionalized with the three different antibodies as the Test Line [8].
| Virus | Source | Bacteria | Source |
|---|---|---|---|
| Adenovirus | BEI, cell preparation | Streptococcus mutans | ATCC |
| Respiratory syncytial virus | ATCC | Streptococcus pneumoniae | ATCC |
| Haemophilus influenzae | Hardy Diagnostics | Streptococcus pyogenes | ATCC |
| Human Metapneumovirus | BEI, Inactive cell lysate | Mycoplasma pneumoniae | ATCC |
| Enterovirus | BEI, cell preparation | Chlamydia pneumoniae | ATCC |
| Rhinovirus | BEI, Inactive cell lysate | Legionella pneumophila | ATCC |
| Influenza A | BEI, Inactive cell lysate | Candida albicans | ATCC |
| Influenza B | BEI, Inactive cell lysate | Mycobacterium tuberculosis | Univ. Rhode Island |
| Human coronavirus 229E | ZeptoMetrix | Interferents | Source |
| Human coronavirus OC43 | ZeptoMetrix | Acetaminophen | Sigma-Aldrich |
| Human coronavirus NL63 | ZeptoMetrix | Amoxicillin | Sigma-Aldrich |
| MERS | BEI, Inactive cell lysate | Aspirin | Sigma-Aldrich |
| SARS-Related Coronavirus 2 | BEI, Heat inactivated (Isolate USA-WA1/2020) |
Caffeine | Sigma-Aldrich |
| Human Blood, EDTA | Lee Biosolutions |
Table 1: Sources for viruses, bacteria, medications, and blood used in this study.
(SARS-CoV-2), isolate USA-WA1/2020, 1.6 × 105 TCID50 per mL |
||||
|---|---|---|---|---|
| Dilution | 1/10 | 1/100 | 1/200 | 1/400 |
| Conc. | 1.6 x 104 | 1.6 x 103 | 8 x 102 | 4 x 102 |
| 5 replicates | 100% (5/5) | 100% (5/5) | 60% (3/5) | 0% (0/5) |
| 20 replicates | 100% (20/20) | 100% (20/20) | 50% (10/20) | 0% (0/20) |
Table 2: COVID-19 saliva antigen at-home test limit of detection data.
Methods
The COVID-19 Saliva Antigen At-Home Test procedure was initially developed by varying the following parameters: sample volume to buffer volume, mixing time, wait time, number of microL added to the cassette and time to detect a visible Test Line. This procedure was performed using 5 purchased SARS-CoV-2 positive saliva samples and 5 pooled, de-identified saliva samples (Lee Bio Solutions), measured by a minimally trained lab technician in a randomized and blind fashion using RTA’s Test kit (Figure 1A). The samples in 1 mL plastic centrifuge tubes were measured according to the following steps: 1) a sterile swab was immersed and swirled in a tube with a sample for 30 seconds (Figure 2A); 2) the swab was then immersed and swirled in a vial containing 0.5 mL extraction buffer for 30 seconds; 3) the swab was pressed along the edge of the vial walls to release the solution into the vial; 4) the vial was capped, shaken, and allowed to sit and incubate for 5 minutes at room temperature; 5) 100 microL of sample were drawn from the vial using an auto-pipetter with disposable tips and dispensed into the sample well of a LFA cassette (Figure 2B); and 6) 10 minutes after deposition, the lab technician was asked if the test was positive or negative (2 lines or 1 line). The lab technician correctly identified the 5 positive and 5 negative sample. Measurements of SARS-CoV-2 on cassettes were initially performed visually, identifing results as positive or negative based on visible lines at the Test and Control Lines or no visible line at the Test Line with a visible line at the Control Line, respectively. After full development of the metod, additional samples were measured using an App (RTA) on a smartphone and a cassette holder to quantify the LFA Test Line Reflectance in terms of a Ct value (Figure 1B).
Figure 2: Photographs of A) the set up to measure “unknown” saliva samples (10 shown) using RTA’s COVID-19 Saliva Antigen Test kit and B) Lab technician measuring the 10th sample. 1) Ten 0.5 mL saliva samples in 1 mL centrifuge tubes, 2) ten vials of 0.5 mL buffer, 3) auto-pipetter, 4) box of pipettes, 5) ten cassettes, and 6) used swabs to be bagged and autoclaved.
Results
LFA performance statistics
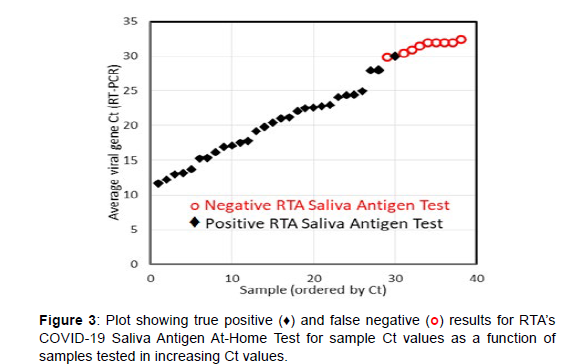
Once the LFA procedure was optimized, the limit of detection (LOD), cross-reactivity, interference, and clinical evaluation were performed. The LOD, also known as the analytical sensitivity, was established by measuring the heat inactivated SARS-Related Coronavirus 2, Isolate USA-WA1/2020, at the concentration of 1.6x105 TCID50 per mL, followed by dilution in pooled, de-identified saliva by factors of 10, 100, 200, and 400 on 5 and 20 replicate LFA cassettes (Table 2). In accordance with FDA, the 100% detection, defined as a visible Test Line for 20 tested samples, yielded an LOD of 1.6x103 TCID50 per mL. The cross-reactivity, also known as the analytical specificity, was established by measuring 12 viruses, which included 3 non-human pathogenic coronaviruses, and 7 bacteria at the purchased concentrations using the LFA cassettes. None of pathogens produced a visible Test Line, confirming no cross-reactivity for these FDA suggested pathogens (Table 3). Similarly, microbial interference was tested by adding a virus, bacteria, biologicals, or drugs to a saliva sample containing SARS-CoV-2 at 1.6x103 TCID50 per mL and measuring each 3 times. None of these FDA suggested potential interferents at relevant concentrations interfered with the expected visible Test Line indicating a positive SARS-CoV-2 detection (Table 4). An evaluation of the LFA cassettes, in terms of true positives, true negatives, false positives and false negatives, was performed by measuring 50 true negative saliva samples and 38 true positive samples. The true negative samples were de-identified pooled saliva, while the true positive samples contained SASR-CoV-2 with Ct values ranging from 11.6 to 32.5 (Lee Biosolutions and Boca Biolistics). All samples were tested with the LFA cassettes 3 times by RTA lab personnel. 100% of the true negative samples did not produce a visible Test Line, while 29 of the 38 true positive samples (76.32%) did produce a visible Test Line. All 9 true positive samples that incorrectly tested negative had Ct values from 30-32.5 (Table 5). Several studies suggested that the ideal antigen test would detect the virus 100% of the time at Ct values below 30, when people are most contagious. The COVID-19 Saliva Antigen At-Home Test met this criterion by identifying all samples with Ct values of 29 and below as positive (Figure 3 and Table 6).
Virus/Bacteria |
Source | Concentration | Result |
|---|---|---|---|
| Adenovirus | BEI, cell preparation | 2.5 x 107 TCID50 /ml | Negative |
| Respiratory syncytial virus | ATCC | 4 x 105 TCID50 /ml | Negative |
| Haemophilus influenzae | Hardy Diagnostics | 3 x 106 TCID50 /ml | Negative |
| Human Metapneumovirus | BEI, Inactive cell lysate | 5 x 105 TCID50 /ml | Negative |
| Enterovirus | BEI, cell preparation | 2.4 x 105 TCID50 /ml | Negative |
| Rhinovirus | BEI, Inactive cell lysate | 2 x 106 TCID50 /ml | Negative |
| Influenza A | BEI, Inactive cell lysate | 6 x 105 CEID50 /ml | Negative |
| Influenza B | BEI, Inactive cell lysate | 5.3 x 104 CEID50 /ml | Negative |
| Human coronavirus 229E | ZeptoMetrix | 1 x 105 TCID50 /ml | Negative |
| Human coronavirus OC43 | ZeptoMetrix | 1 x 105 TCID50 /ml | Negative |
| Human coronavirus NL63 | ZeptoMetrix | 1 x 105 TCID50 /ml | Negative |
| MERS | BEI, Inactive cell lysate | 8.9 x 105 TCID50 /ml | Negative |
| Streptococcus pneumoniae | ATCC | 5 x 106 cells /ml | Negative |
| Streptococcus pyogenes | ATCC | 8 x 105 cells /ml | Negative |
| Mycoplasma pneumoniae | ATCC | 3.2 x 106 cells /ml | Negative |
| Chlamydia pneumoniae | ATCC | 7.5 x 107 cells /ml | Negative |
| Legionella pneumophila | ATCC | 5 x 105 cells /ml | Negative |
| Mycobacterium tuberculosis | Univ. Rhode Island | 6.3 x 106 cells /ml | Negative |
| Candida albicans | ATCC | 4 x 106 cells /ml | Negative |
Table 3: COVID-19 saliva antigen at-home test cross-reactivity data.
TCID50 = 50% Tissue Culture Infectious Dose, CEID50 = 50% Chicken Embryo Infectious Dose
Interferent |
Concentration | SARS-CoV-2 Concentration | Result, n = 3 |
|---|---|---|---|
| Influenza A | 6x105 CEID50 /ml | 1.6x103 TCID50 per mL | Positive |
| Streptococcus pneumoniae | 5x106 cells /ml | 1.6x103 TCID50 per mL | Positive |
| Streptococcus mutans | 1x107 cells /ml | 1.6x103 TCID50 per mL | Positive |
| Human Blood, EDTA | 5% v/v | 1.6x103 TCID50 per mL | Positive |
| Acetaminophen | 1 mg/ml | 1.6x103 TCID50 per mL | Positive |
| Aspirin | 1mg/ml | 1.6x103 TCID50 per mL | Positive |
| Caffeine | 5mg/ml | 1.6x103 TCID50 per mL | Positive |
| Amoxicillin | 5 mg/ml | 1.6x103 TCID50 per mL | Positive |
Table 4: COVID-19 Saliva antigen at-home test interferent data.
LFA Statistics |
Sensitivity | Specificity | Predicted Positive | Predicted Negative |
|---|---|---|---|---|
| a/(a+c) | d/(d+b) | a/(a+b) | d/(d+c) | |
| Ct =13-33 | 29/(29+9) | 50/(50+0) | 29/(29+0) | 50/(50+9) |
| a=29, b=0, c=9, d=50 | 76.32% | 100.00% | 100.00% | 84.75% |
| Ct =13-29 | 29/(29+0) | 50/(50+0) | 29/(29+0) | 50/(50+0) |
| a=29, b=0, c=0, d=50 | 100.00% | 100.00% | 100.00% | 100.00% |
Table 5: COVID-19 saliva antigen at-home test sensitivity, specificity, predicted positive, and
predicted negative percents are provided for Ct values 11.6 to 32.5 and 11.6 to 30.
Positive Samples |
PCR Ct value | LFA Test | Positive Samples | PCR Ct value | LFA Test |
|---|---|---|---|---|---|
| 1 | 21.2 | Positive | 20 | 13.1 | Positive |
| 2 | 22.5 | Positive | 21 | 15.3 | Positive |
| 3 | 19.8 | Positive | 22 | 16.9 | Positive |
| 4 | 23 | Positive | 23 | 20.4 | Positive |
| 5 | 16.2 | Positive | 24 | 22.8 | Positive |
| 6 | 22.6 | Positive | 25 | 24.1 | Positive |
| 7 | 12.2 | Positive | 26 | 31.5 | Negative |
| 8 | 17.1 | Positive | 27 | 30.5 | Negative |
| 9 | 24.5 | Positive | 28 | 32.5 | Negative |
| 10 | 13.7 | Positive | 29 | 28 | Positive |
| 11 | 15.3 | Positive | 30 | 25 | Positive |
| 12 | 32 | Negative | 31 | 13 | Positive |
| 13 | 19.2 | Positive | 32 | 21 | Positive |
| 14 | 24.4 | Positive | 33 | 32 | Negative |
| 15 | 22.1 | Positive | 34 | 30 | Positive |
| 16 | 30 | Negative | 35 | 28 | Positive |
| 17 | 17.5 | Positive | 36 | 31 | Negative |
| 18 | 17.7 | Positive | 37 | 32 | Negative |
| 19 | 11.6 | Positive | 38 | 32 | Negative |
Table 6: List of true positive and false negative results for 38 samples as determined by RTA’s COVID-19 saliva antigen at-home test (actual Ct values shown).
LFA Quantitation
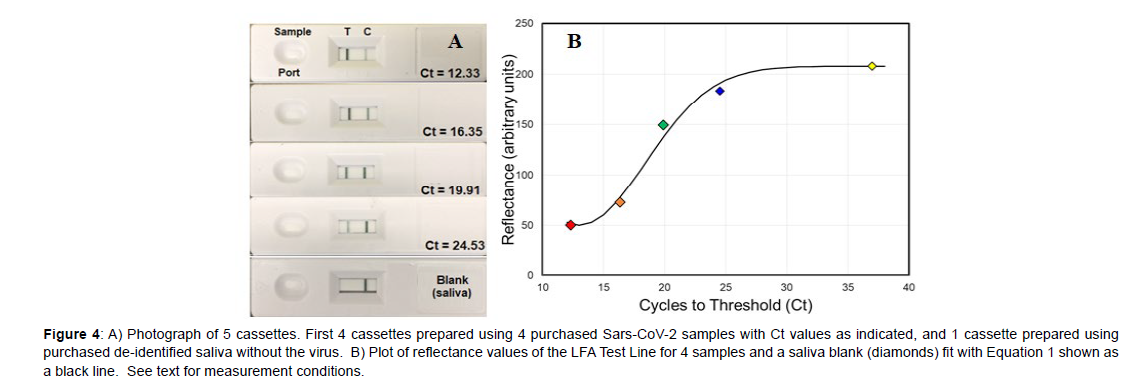
While analyzing the 38 samples described above it became clear that the Test Line intensity decreased with lower viral loads, i.e. increasing Ct values. In an effort to quantify this intensity several spectroscopic methods were investigated, such as surface-enhanced Raman spectroscopy and both transmission and reflectance visible spectroscopy [8]. Since reflectance showed an inverse relationship to the Test Line intensity, measurement of reflectance using a smartphone was tried, as it would be ideal for at-home tests. During preliminary measurements it was found that a stand was required to fix the smartphone camera above the cassette Test Line for reproducible alignment (Figure 1B). A basic smartphone App was written using the built in camera to 1) magnify the Test Line image, and 2) measure the Test Line reflectance. Four saliva samples covering most of the Ct range, specifically Ct values of 12.33, 16.35, 19.91, and 24.53, were purchased, and the reflectance measured to establish a correspondence to the Ct values. The samples were prepared and added to cassettes as described previously. For each sample, the reflectance was measured at 10 minutes after it was added to the cassette sample port using the smartphone camera. In addition, a saliva sample, devoid of SARS-CoV-2, was also measured to represent a Test Line containing no probes and the upper limit of reflectance.
The Ct 12.33 was used to represent a Test Line saturated with probes and the lower limit of reflectance. The Test Line reflectances for these two samples were quantified using the smartphone App as 208 and 50, respectively. There is a clear decrease in visible intensity of the Test Lines for the 5 samples as the viral load decreases (Figure 4A). A plot of the reflectance as a function of Ct values for the 4 samples and the blank saliva sample was fit with the Avrami equation (Figure 4B, Equation 1) [9], used to describe a limited system that has initial exponential growth followed by exponential decay. For the present system, an exponential increase in antibody binding is expected at the Test Line as the sample concentration increases, followed by an exponential decrease as all the bonding sites become occupied.
Figure 4: A) Photograph of 5 cassettes. First 4 cassettes prepared using 4 purchased Sars-CoV-2 samples with Ct values as indicated, and 1 cassette prepared using purchased de-identified saliva without the virus. B) Plot of reflectance values of the LFA Test Line for 4 samples and a saliva blank (diamonds) fit with Equation 1 shown as a black line. See text for measurement conditions.
Ct = 208-157(exp[-0.014x(Reflectance-12)2]) (1)
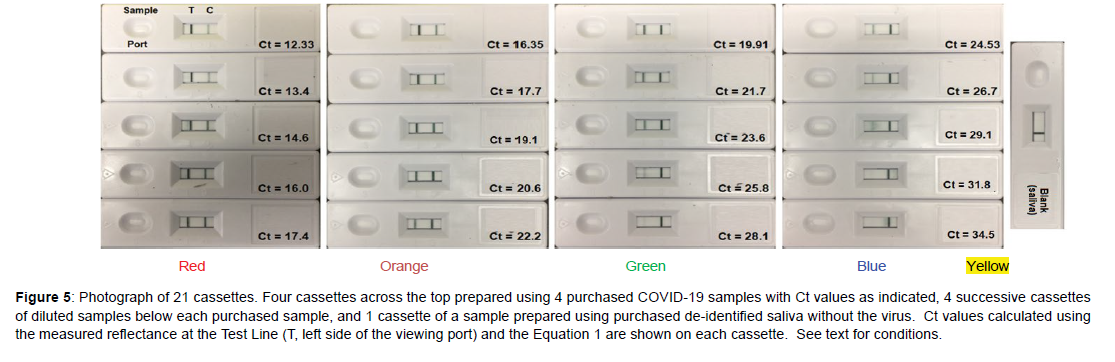
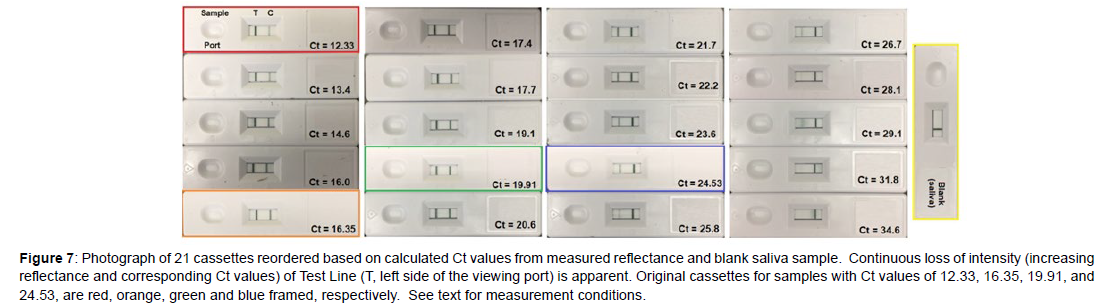
Next, a sample set of 16 “unknowns” were prepared by diluting each of the 4 purchased samples 4 times in de-identified, pooled saliva by 50% (Figure 5). This dilution factor was chosen to mimic the factor of 2 nucleic acid replication achieved from one PCR cycle to the next. As before, the samples were prepared, added to cassettes, and the reflectance measured at the Test Line, but this time the smartphone was also used to calculate the Ct values using Equation 1. However, the calculated Ct values did not match the Ct values based on 50% dilution (Figure 6A, Table 7). Notably, the intensity of the purchased Ct sample with a value of 19.91 should equal 23.91 after 4 dilutions, but its Test Line is lighter than the purchased sample with a Ct value of 24.53. In an effort to correct this discrepancy, the Ct gaps between the original samples were used to correct all of the diluted sample Ct values by a multiplication factor of 1.086 (Figure 6B and Table 7). The correction factor indicates that the replication for each cycle was not a factor of 2.0, but 1.84 (2.0/1.086). This is not unusual, as polymerase chain reaction replications are not 100% efficient, as occasional sequencing errors occur during each cycle [10, 11]. Reordering the sample cassettes based on this correction shows a continual visible decrease in the Test Line intensity from the Ct 12.33 sample to the blank saliva sample (Figure 7).
Figure 5: Photograph of 21 cassettes. Four cassettes across the top prepared using 4 purchased COVID-19 samples with Ct values as indicated, 4 successive cassettes of diluted samples below each purchased sample, and 1 cassette of a sample prepared using purchased de-identified saliva without the virus. Ct values calculated using the measured reflectance at the Test Line (T, left side of the viewing port) and the Equation 1 are shown on each cassette. See text for conditions.
Figure 6A: Plot of reflectance values of the LFA Test Line for the 16 diluted samples calculated using Equation 1. Diluted samples are represented as red, orange, green, and blue circles, prepared from Ct values of 12.33, 16.35, 19.91, and 24.53, respectively. B) Plot of the same samples, but the Ct values corrected by multiplying by 1.086. Equation 1 is shown as a black line in both figures. See text for measurement conditions.
Figure 7: Photograph of 21 cassettes reordered based on calculated Ct values from measured reflectance and blank saliva sample. Continuous loss of intensity (increasing reflectance and corresponding Ct values) of Test Line (T, left side of the viewing port) is apparent. Original cassettes for samples with Ct values of 12.33, 16.35, 19.91, and 24.53, are red, orange, green and blue framed, respectively. See text for measurement conditions.
Sample |
Prepared | Eq 1 Predicted | Measured | Corrected | Calculated | Ct Error |
|---|---|---|---|---|---|---|
| Number | Ct Values | Reflectance | Reflectance | Ct Values | Ct Values | Corr-Calc |
| 1 | 12.326 | 51.22 | 50 | 12.3 | 12.3 | |
| 2 | 13.326 | 50.29 | 52.33 | 13.4 | 14.1 | -0.7 |
| 3 | 14.326 | 54.65 | 64.33 | 14.5 | 15.2 | -0.7 |
| 4 | 15.326 | 63.88 | 78 | 15.8 | 16.3 | -0.5 |
| 5 | 16.326 | 77.09 | 83 | 17.1 | 16.8 | 0.3 |
| 6 | 16.35 | 77.44 | 72.67 | 16.4 | 16.4 | |
| 7 | 17.35 | 93.46 | 93.33 | 17.8 | 17.5 | 0.3 |
| 8 | 18.35 | 110.87 | 121 | 19.3 | 19 | 0.3 |
| 9 | 19.35 | 128.39 | 154 | 20.9 | 20.9 | 0 |
| 10 | 20.35 | 144.93 | 166 | 22.7 | 21.9 | 0.8 |
| 11 | 19.91 | 137.83 | 149.33 | 19.9 | 19.9 | |
| 12 | 20.91 | 153.31 | 171 | 21.6 | 22.3 | -0.7 |
| 13 | 21.91 | 166.9 | 182.33 | 23.5 | 23.5 | 0 |
| 14 | 22.91 | 178.14 | 200 | 25.5 | 27 | -1.5 |
| 15 | 23.91 | 187.04 | 207.67 | 27.7 | 35 | -7.3 |
| 16 | 24.53 | 191.51 | 183.33 | 24.5 | 24.5 | |
| 17 | 25.53 | 197.05 | 197.33 | 26.6 | 25.6 | 1 |
| 18 | 26.53 | 200.97 | 200.67 | 28.9 | 26.6 | 2.3 |
| 19 | 27.53 | 203.64 | 204.67 | 31.4 | 29 | 2.4 |
| 20 | 28.53 | 205.38 | 206.67 | 34.1 | 30 | 4.1 |
| 21 | 37 | 208 | 208 | 37.1 | Ave Error | 1.4 |
| Std Dev | 1.9 |
Table 7: Comparisons of Ct values for prepared samples by dilution, predicted reflectance for prepared samples, and measured reflection for prepared samples. Corrected predicted Ct values (x1.086), Calculated Ct values based on Equation 1, and Ct error (Corrected minus Calculated Ct values) are listed. Average error (absolute) and standard deviation between these values are also provided.
Discussion
The described saliva-based at-home SARS-CoV-2 test met the FDA EUA requirements for sensitivity, cross-reactivity, and interference using purchased saliva samples containing the virus at known Ct values. Furthermore, the statistical measurements of sensitivity, specificity, predicted positive and predicted negative were all 100% correct for samples with Ct values of 29 and below. However, a clinical study was not performed using the kit, which would have included 1) collecting a saliva sample using a swab or passive drool, 2) mixing the saliva sample with buffer, 3) transferring the sample to a cassette, and 4) visually determining if a line appeared or not at the Test Line between 15 and 20 minutes. A preliminary laboratory study of saliva samples with known Ct values ranging from 12 to 38 Ct values was successfully used to establish a relationship between the smartphone measured reflectance at the LFA Test Line and the Ct values. However, as presented in Table 7, there remains a minor discrepancy between the corrected Ct values and the calculated Ct values with an average difference of 1.4. There are three potential sources of error, 1) preparation of the diluted samples, 2) the accuracy at which the smartphone camera is aligned with the Test Line, and 3) the probe to antibody binding distribution at the Test Line. Nevertheless, this error is quite small in terms of making decisions regarding the level of a person’s infection and contagious levels. In contrast, the standard deviation of 1.9 is somewhat high, although it is skewed by Sample 15, the fourth dilution of the Ct 19.9 sample, with a “corrected minus calculated Ct value” of -7.3. Removing this one sample improves both the average error and standard deviation to 1.05±1.12. Furthermore, the error increases significantly above Ct 26 as the reflectance approaches 200, 95% of the measureable reflectance limit of 208 ([208-200]/[208-50]). Eliminating Samples 15, 18, 19, and 20 yields an average difference and standard deviation of 0.58±0.43 for the more critical diagnostic regions: highly contagious, Ct < 21, and contagious, Ct 21 to Ct 27 [12].
Conclusion
A simple four step at-home SARS-CoV-2 test was developed and successfully used to perform FDA EUA required laboratory measurements for lateral flow assay based kits. The test employed saliva, which could prove useful for testing children. The test also employed three antibodies to bind to the virus, which provided excellent sensitivity and selectivity, and should also allow detecting future variants. The ability of measuring the reflectance of the LFA Test Line for a saliva sample using a smartphone to calculate Ct values, and estimate the level of SARS-CoV-2 infection was also demonstrated. The relationship between the measure reflectance and Ct values followed the Avrami equation, which has been previously demonstrated for PCR plots of sample fluorescence as a function of Ct values [13]. While the upper Ct limit of 26-27 was determined for quantitative measurements, an upper limit for true positives was found to be 29-31 for 38 purchased saliva samples. The small scatter in the data from the Avrami curve demonstrates that the performance of the LFAs were very consistent in terms of probe-to-antigen binding at the conjugate pad and antigento- antibody binding at the Test Line, as well as flow across the LFAs. The consistency and quantitative nature of the data also suggests the LFAs could possibly be used to indicate the level of infection [12-16]. Furthermore, the use of the smartphone App and LFAs could be used at-home by individuals to estimate their infection and contagious levels, self-isolate as necessary, and anonymously share their data with health agencies, such as the CDC, to better track and limit outbreaks. The combination lateral flow assay and smartphone quantitation is well suited for identifying, quantifying, and quarantining to slow the spread of future viruses.
Acknowledgement
Knowledge regarding the design and development of the probes and the LFAs was learned during a Department of Defense, USA Medical Research program (W81XWH19-C-0079).
References
- (2022) www.worldometers.info/coronavirus/.
- Cumbers J (2020) “One company was the first to produce a key component of the Covid-19 PCR test. And it has now provided 52 million tests” Forbes.
- Barber G (Mar 16, 2020) FDA approves the first commercial corona virus tests in the US, Wired-Science.
- Monitoring and tracking the disease (July 1, 2020) Centers for Disease Control and Prevention.
- Larremore DB, Wilder B, Lester E, Shehata S, Burke JM, et al. (2021) Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Science advances 7:eabd5393.
- COVID-19 (July 1, 2020) World Health Organization.
- Farquharson S, Shende C (2022) “Detection of Tacrolimus in Saliva using a Lateral Flow Assay and Surface-Enhanced Resonance Raman Scattering”. J Anal Bioanal Tech 13.
- Shende C, Farquharson D, Farquharson S(2023)“Quantitation of Chemical, Biochemical, and Biological analytes on a flow assay using a smartphone submitted US Patent Office.
- Melvin A (1939) Kinetics of phase change. I General theory. The Journal of Chemical Physics 7: 1103-1112.
- Vogels CB, Brito AF, Wyllie AL, Fauver JR, Ott IM, et al. (2020) Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nature microbiology 5:1299-1305.
- Valley-Omar Z, Marais G, Iranzadeh A, Naidoo M, Korsman S, et al. (2022) Reduced amplification efficiency of the RNA-dependent-RNA-polymerase target enables tracking of the Delta SARS-CoV-2 variant using routine diagnostic tests. Journal of Virological Methods 302:114471.
- Mina MJ, Parker R, Larremore DB (2020) Rethinking Covid-19 test sensitivity—a strategy for containment. New England Journal of Medicine 383:e120.
- Victoriano CM, Pask ME, Malofsky NA, Seegmiller A, Simmons S, et al. (2022) Direct PCR with the CDC 2019 SARS-CoV-2 assay: optimization for limited-resource settings. Scientific Reports 12:1-12.
- Singanayagam A, Patel M, Charlett A, Bernal JL, Saliba V, et al. (2020) Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Eurosurveillance 25:2001483.
- Rabaan AA, Tirupathi R, Sule AA, Aldali J, Mutair AA, et al. (2021) Viral dynamics and real-time RT-PCR Ct values correlation with disease severity in COVID-19. Diagnostics 11:1091.
- Jones TC, Biele G, Mühlemann B, Veith T, Schneider J, et al. (2021) Estimating infectiousness throughout SARS-CoV-2 infection course. Science 373: eabi5273.
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Citation: Shende C, Farquharson D, Farquharson S (2023) Development of aSaliva-Based Lateral Flow Assay for SARS-Cov-2 with the Potential to QuantifyViral Load. J Anal Bioanal Tech 14: 536.
Copyright: © 2023 Shende C, et al. This is an open-access article distributed underthe terms of the Creative Commons Attribution License, which permits unrestricteduse, distribution, and reproduction in any medium, provided the original author andsource are credited.
Share This Article
Open Access Journals
Article Usage
- Total views: 1273
- [From(publication date): 0-2023 - Apr 04, 2025]
- Breakdown by view type
- HTML page views: 1054
- PDF downloads: 219