Developing a PANoptosis Signature: Identification of Unique Immunotherapeutic Candidates for Osteosarcoma
Received: 10-Jul-2024 / Manuscript No. JIDT-24-141283 / Editor assigned: 12-Jul-2024 / PreQC No. JIDT-24-141283 (PQ) / Reviewed: 26-Jul-2024 / QC No. JIDT-24-141283 / Revised: 02-Aug-2024 / Manuscript No. JIDT-24-141283 (R) / Published Date: 09-Aug-2024 DOI: 10.4172/2332-0877.1000598
Abstract
This study focused on elucidating the role of PANoptosis in Osteosarcoma (OS), a highly malignant bone tumor. By screening and integrating OS-related microarray datasets from Gene Expression Omnibus (GEO), we identified 105 PANoptosis-related differentially expressed genes (OS_PAN_DEGs) primarily involved in apoptosis, necroptosis, proteasome, hippo signaling, and neurodegenerative disease pathways. These genes were used to classify OS into three distinct subtypes with varying clinical outcomes, immune characteristics, and mutational landscapes. Additionally, we developed an OS_PAN index model to assess the association between PANoptosis and OS features, treatment response, and prognosis. Notably, high OS_PAN-index patients responded well to immunotherapy, while low-index patients showed sensitivity to small-molecule targeted drugs. Drug screening revealed Pazopanib, Chelerythrine, Staurosporine, Hydroxyurea, and Sunitinib as potential therapeutic agents positively correlated with OS_PAN_DEGs expression. This comprehensive analysis enhances our understanding of OS pathogenesis and offers novel therapeutic targets for OS treatment.
Keywords: Osteosarcoma (OS); PANoptosis; Immune characteristics; Drug sensitivity; Prognostic index
Introduction
Osteosarcoma (OS) is a malignant bone tumor that primarily affects children, adolescents, and young adults [1]. Its clinical manifestations are diverse, with common symptoms including pain, mass, limited mobility, and pathological fractures [2]. In recent years, OS, as a malignant tumor, has garnered widespread attention globally. Despite certain progress made in clinical treatment, the incidence and mortality rates of OS remain high, severely affecting the quality of life and lifespan of patients [3].
The primary treatment modalities for OS mainly include surgical resection, radiotherapy, and chemotherapy, among other traditional therapeutic methods [4]. However, these approaches have certain limitations. Surgical resection can be effective for early-stage OS, but, for advanced cases, the surgical difficulty is high, postoperative recovery is slow, and it can easily lead to functional impairment [5]. The control effect of radiotherapy on OS is limited, and the side effects of radiotherapy are considerable, easily damaging surrounding normal tissues [6]. Chemotherapy also has certain restrictions in the treatment of OS, as some patients cannot tolerate it due to drug resistance or adverse reactions [7]. Therefore, it is particularly important to search for new therapeutic targets and strategies.
The characteristics of PANoptosis, also known as non-apoptotic cell death, have become a popular research direction in the field of immunotherapy in recent years. PANoptosis is a form of cell death distinct from the classical apoptotic death pathway, exhibiting different morphological manifestations and regulatory mechanisms [8]. PANoptosis characteristics have been proven to play a major role in tumor genesis and development [9]. Firstly, the interaction between PANoptosis characteristics and tumor cells can be achieved through apoptotic pathways [10]. The emergence of PANoptosis characteristics can restore the function of abnormally expressed apoptosis-related proteins, guiding tumor cells to re-enter the apoptotic pathway and eliminate them [11]. Secondly, PANoptosis characteristics can increase the expression of tumor antigens, activate immune cells, and enhance their ability to kill tumor cells [12,13]. Finally, the expression of PANoptosis characteristics can also regulate the invasion and metastasis abilities of tumor cells, inhibiting tumor progression and metastasis [14,15]. However, there is still relatively limited research on its role and mechanisms in the immunotherapy of OS.
In recent years, immunotherapy, as a novel therapeutic approach, has gradually become a research hotspot and attracted much attention from researchers [16]. Immunotherapy aims to treat tumors by enhancing the patient’s own immunity and regulating their immune system to recognize and attack cancer cells [17,18]. In the immunotherapy of OS, the current main methods include immune checkpoint inhibitors, cellular immunotherapy, and vaccine therapy [19,20]. However, despite some important progress made in the field of immunotherapy, there are still some issues that need to be addressed. On the one hand, OS exhibits a certain degree of heterogeneity, and there are differences among different patients, leading to varying responses to immunotherapy [21]. On the other hand, OS cells possess certain immune evasion mechanisms, such as expressing inhibitory costimulatory molecules that suppress the activation and killing functions of immune cells, thereby reducing the effectiveness of immunotherapy [22,23]. Therefore, a deeper understanding of the mechanisms of PANoptosis can provide new opportunities for developing effective immunotherapy strategies for OS. Currently, there is a gap in research on the role of PANoptosis in OS.
In this study, we employed consensus-cluster-plus analysis to identify three subpopulations based on differentially expressed genes (OS_PAN_DEGs) related to PANoptosis and OS. Subsequently, we investigated the immune characteristics and mutational landscapes of these subpopulations and constructed a PANoptosis risk scoring model (OS_PAN-index). Utilizing this PANoptosis signature, we evaluated the potential association between PANoptosis and OS characteristics, as well as its predictive value for treatment response and prognosis.
Our findings may pave the way for innovative targeted therapies in the treatment of OS patients.
Materials and Methods
Data resources
Microarray datasets related to OS were downloaded from the national center for biotechnology information Gene Expression Omnibus (GEO) database [24]. The screening criteria were as follows: The microarray dataset should be a gene expression profile; the dataset should be a gene expression profile related to OS tissue samples; OS samples should be derived from primary and metastatic sites; the gene expression profile should be from human OS; and the total number of OS samples should be greater than 20. Datasets that did not meet any of the criteria were excluded. Finally, three datasets were selected for further analysis: GSE16088, GSE21257 and GSE14359 [25-27]. Among them, GSE16088 and GSE14359 datasets were used to screen for differentially expressed genes. Initially, the datasets were merged using the R package in silico merging, and batch effects were removed using the remove batch effect function from the R package limma. This resulted in a matrix with batch effects removed. The GSE21257 database was used as the training set, including 53 OS patients and their clinical information (age, gender, histological subtype and huvos grade, tumor location, metastasis at diagnosis and survival information). The validation set, Therapeutically Applicable Research to Generate Effective Treatment-Osteosarcoma (TARGET-OS) dataset (n=95), was obtained from the TARGET database. Detailed information on the datasets is provided in Table 1.
| Dataset/Cohort | Database | Data type | Sample information |
|---|---|---|---|
| GSE16088 | GEO | RNA-seq data | 14 cases of osteosarcoma tissue samples + 6 cases of normal tissue samples |
| GSE14359 | GEO | RNA-seq data | 10 cases of osteosarcoma tissue samples + 2 cases of normal tissue samples |
| GSE21257 | GEO | RNA-seq data + clinical information | 53 cases of osteosarcoma tissue samples |
| TARGET-OS | TARGET | RNA-seq data + clinical information | 95 cases of osteosarcoma tissue samples |
Table 1: Data source
| Drug | DsigDB.p-value | Cellminer.p-value | Cellminer.Cor | Genes |
|---|---|---|---|---|
| Pazopanib | 0.009561 | 0.007298 | 0.343007 | IL6 |
| Chelerythrine | 0.013522 | 0.012157 | 0.321832 | IL6; CYCS |
| Elesclomol | 0.019829 | 0.000108 | -0.47892 | BTG3; PSME4 |
| Staurosporine | 3.06E-04 | 0.012342 | 0.321183 | IL6 ;CYCS |
| Tamoxifen | 0.002599 | 0.000151 | -0.46998 | IL6; CYCS; BTG3 |
| Hydroxyurea | 0.037767 | 0.012742 | 0.319809 | BTG3; CYCS |
| Fulvestrant | 1.95E-04 | 0.000965 | -0.41541 | IL6; PSME4; CYCS; BTG3 |
| Sunitinib | 0.016286 | 0.013166 | 0.31839 | IL6; STAT6 |
Table 2: Drug screening
We collected five pathways downloaded from the Molecular Signatures Database (MSigDB) (version 7.4) (Gene Set Enrichment Analysis |MSigDB: gsea-msigdb.org) for analysis, including reactome_ pyroptosis, hallmark_apoptosis, Kyoto Encyclopedia of Genes and Genomes (KEGG)_apoptosis, reactome_apoptosis, and map04217 pathway (KEGG_necroptosis) [28]. The union of all gene sets from these five pathways was designated as the PANoptosis pathway gene set (Supplementary Table 1).
Identification of OS-related gene modules
The Weighted Gene Co-Expression Network Analysis (WGCNA) method was utilized to identify gene modules associated with OS. WGCNA not only provides information on the correlation between two nodal genes and their related genes, but also offers topological properties of co-expression networks [29]. The WGCNA package was employed in R3.4.1 software to screen for stable gene modules in the merged dataset of GSE16088 and GSE14359.
Identification of Differentially Expressed Genes (DEGs)
Firstly, the raw datasets of GSE16088 and GSE14359 were background-corrected, normalized and log2-transformed using the affy package in R. When multiple probes identified the same gene, their average expression values were calculated to determine its expression level. After merging these two datasets, the bioconductors “Surrogate Variable Analysis (SVA)” R package was applied to eliminate batch effects. Differential gene analysis was performed using the “limma” package in R with a cut-off of P2 to distinguish DEGs between disease and healthy samples in the merged dataset. Subsequently, volcano plots and heatmaps were employed to visualize the DEG expression data [30].
Unsupervised cluster analysis of OS-PANoptosis-related model genes
We performed consensus clustering analysis using the “consensus- cluster-plus” package and model genes to identify unknown OS subtypes [31]. The clustering process utilized 1-Pearson correlation distance, with a sample resampling rate of 80%, and was repeated ten times. We employed the empirical cumulative distribution function plot to determine the optimal number of clusters.
Functional enrichment analysis
To perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis, we utilized the R packages “org.Hs.eg.db” and “clusterProfiler” (version 3.14.3). Firstly, we annotated the genes with GO terms using “org.Hs.eg.db” and mapped them to the background set. Subsequently, we employed the “clusterProfiler” package to conduct GO and KEGG enrichment analysis and obtain the gene set enrichment results. In both cases, the minimum and maximum sizes of the gene sets were set to 5 and 5000, respectively. We retrieved the latest KEGG pathway gene annotations through the KEGG rest api and mapped them to the background set. The statistical significance of both analyses was determined by a P-value [32].
For Gene Set Enrichment Analysis (GSEA), we obtained subset collections from the molecular signatures database to assess relevant pathways and molecular mechanisms based on gene expression profiles and phenotypic groupings [33]. We performed 1000 permutations to obtain statistically significant results with a P-value
Analysis of somatic mutations
To assess somatic mutations and Tumor Mutation Burden (TMB), we utilized the R package “maftools” [34,35]. Somatic mutation data were obtained from the TARGET database and analyzed to identify nonsynonymous somatic mutations. Subsequently, the TMB score was calculated by dividing the number of non-synonymous somatic mutations by the total size of the genome in megabases.
The Tumor Immune Dysfunction and Exclusion (TIDE) framework is a computational tool that utilizes gene expression profiles of cancer samples to assess the potential for tumor immune evasion [36,37]. The TIDE score, calculated for each tumor sample, serves as a biomarker to predict the response to immune checkpoint blockade, including Anti-Programmed Cell Death -1 (Anti-PD1) and Anti Cytotoxic T-Lymphocyte-Associated Antigen 4 (Anti-CTLA4), across different cancer types. We employed five algorithms to evaluate immune cell infiltration in the tumor microenvironment: Timer, epic, xcell, cibersort, and mcpcount [38,39] . These algorithms provide a comprehensive assessment of the immune cell landscape within the tumor microenvironment.
Survival analysis and machine learnings
We employed the “glmnet” package to establish a Lasso regression model and utilized 10-fold cross validation to select the optimal lambda value, aiming to enhance the interpretability and predictive accuracy of the model. We determined the coefficient for each gene through multivariate Cox analysis and generated the final regression model using the selected lambda value.
Initially, patients were divided into two groups based on risk coefficient values using the percentile (50%) approach, categorizing them as either high OS_PAN index or low OS_PAN index groups. Subsequently, we used the “survfit” function from the “survival” R package to analyze the prognostic differences between the two groups. The log-rank test method was adopted to assess the significance of prognostic differences between samples in different groups.
Drug screening based on core genes
To facilitate the development of therapeutic approaches, we explored potential drug molecules significantly associated with core genes utilizing the Drug Signature Database (DSigDB) and cell miner database [40,41].
Statistical analysis
Statistical analysis was performed using R software (v.4.1.0). Continuous variables were expressed as mean ± Standard Deviation (SD), while categorical variables were represented as frequencies (percentages). For continuous variables, student’s t-test or Wilcoxon test was used to test differences between two groups based on the assumption of normality of the data. For categorical variables, chisquare test or fisher’s exact test was employed based on the expected frequency counts. In all tests, a two-sided P-value less than 0.05 was considered statistically significant. Kaplan-meier method was adopted for survival assessment, and the log-rank test was used to compare differences between groups. Multivariate survival analysis was performed using the Cox proportional hazards regression model.
Results
Dataset evaluation
The combined gene expression levels of the GEO series after batch effect correction were standardized, and the results before and after standardization are shown in Figures 1A-1D. In the GSE14359 and GSE16088 datasets, we identified probes corresponding to 13,231 genes and confirmed the DEGs in OS. After filtering, a total of 13,231 molecules were obtained, among which 2,835 genes met the threshold of |log2 (FC)| ≥ 1 and p.adj< 0.05 (Figures 1E and 1F).
Figure 1: Identification of differentially expressed genes in the merged osteosarcoma dataset. Based on the cut-off criteria: Absolute |log2FC|>1 and P value<0.05. (A): Box plot of gene expression before normalization in two selected GEO (Gene Expression Omnibus) databases; (B): Box plot of gene expression after normalization in two selected GEO databases; (C): Umap distribution plot before batch effect removal in two selected GEO databases; (D): Umap distribution plot after batch effect removal in two selected GEO databases; (E): Volcano plot of differentially expressed genes after merging two selected GEO datasets; (F): Clustered heat map of differentially expressed genes after merging two selected GEO datasets. 
Weighted Gene Co-expression Network Analysis (WGCNA) and identification of key modules
Here, we employed WGCNA to identify modules most relevant to OS. Based on scale-free topology and mean connectivity, we selected β=14 (scale-free R2=0.82) as the “soft” threshold (Figure 2A). Figure 2B depicts the clustering dendrogram for OS and control groups. Using this power value, we generated 11 Gene Co-Expression Modules (GCMs), presented in Figure 2C with distinct colors. The correlation between OS and GCMs is shown in Figures 2D-2F, where the turquoise module (consisting of 3136 genes), grey module (consisting of 1461 genes), and darkolivegreen module (consisting of 3628 genes) exhibited the highest correlation with OS (turquoise: r=-0.87, p=7e -11; grey: R=0.76, p = 4e- 07; dark olive green: R= 0.62, p=1e-04), and thus were considered as key modules for subsequent analysis.
Figure 2: WGCNA Co-expression and enrichment analysis in Osteosarcoma (OS) Patients. (A): Network topology analysis under different soft-thresholding powers; (B): Cluster dendrogram of expressed genes in OS; (C): Module-trait relationships in OS. Each cell contains the corresponding correlation and p-value; (D): Correlation between genes in the turquoise module and OS; €: Correlation between genes in the grey 60 module and OS; (F): Correlation between genes in the dark olive green module and OS.
Identification and functional analysis of PANoptosis related differentially expressed genes in OS
The PANoptosis gene set encompassed genes related to pyroptosis (27 genes), apoptosis (298 genes) and necroptosis (160 genes) (Figure 3A). The DEGs in OS included 2835 DEGs screened from the GEO database and 8225 genes from the modules most relevant to OS identified by WGCNA (Figures 1C and 1D, and (Supplementary Figure S1). By mapping the three gene sets screened from GEO, WGCNA, and PANoptosis, we obtained 105 genes, which were defined as OS differentially expressed genes (OS_ PAN_DEGs) related to PANoptosis (Figure 3B). We performed GO, KEGG and GSEA enrichment analyses on the OS_PAN_DEGs to explore their biological functions and related signaling pathways.
The Biological Process (BP) analysis revealed that the OS_PAN_ DEGs were mainly enriched in processes such as cellular metabolic regulation, hematopoietic stem cell differentiation, interleukin-1- mediated signaling, and RNA transcription regulation (FDR< 0.1, p-value< 0.05), (Figure 3C). The Molecular Function (MF) analysis demonstrated that the OS_PAN_DEGs were primarily enriched in protein binding, enzyme binding, enzyme regulator activity, and drug binding (FDR<0.1, p-value<0.05), (Figure 3D). The Cellular Component (CC) analysis showed that the OS_PAN_DEGs were mainly enriched in proteasome complexes, endopeptidase complexes, peptidase complexes, and the cytoplasm (FDR<0.1, p-value<0.05), (Figure 3E). KEGG enrichment analysis indicated that the OS_PAN_DEGs were primarily enriched in apoptosis, necroptosis, proteasome, hippo signaling pathway and multiple pathways related to neurodegenerative diseases (FDR<0.1, p-value<0.05), (Figure 3F).
Figure 3: Identification of PANoptosis-related differential genes in Osteosarcoma (OS) and GO/KEGG/GSEA enrichment analysis of OS_PAN_DEGs.(A): PANoptosis gene list; (B): Venn diagram composed of GEO differential genes, WCGNA module genes, and PANoptosis-related datasets; (C-E): GO enrichment analysis based on OS_PAN_DEGs; (F): KEGG enrichment analysis based on OS_PAN_DEGs; G–I: GSEA analysis based on Kyoto Encyclopedia of Genes and Genomes (KEGG), hallmark, and reactome datasets, respectively.
Note:  APOPTOSIS(ES=0.3519,NP=0.0471), MTORC1_SIGNALINING (ES=0.5467,NP=0.0125), SIGNALINING_BY_VEGF(ES-0.4987,NP=0.0396);
APOPTOSIS(ES=0.3519,NP=0.0471), MTORC1_SIGNALINING (ES=0.5467,NP=0.0125), SIGNALINING_BY_VEGF(ES-0.4987,NP=0.0396);  FOCAL_ADHESION (ES=0.6467, NP=0.0224), ALLOGRAFT_REJECTIONS (ES=0.5157, NP=0.0385), CELL_CYCLE (ES=0.4000, NP=0.0483);
FOCAL_ADHESION (ES=0.6467, NP=0.0224), ALLOGRAFT_REJECTIONS (ES=0.5157, NP=0.0385), CELL_CYCLE (ES=0.4000, NP=0.0483);  ALZHEIMERS_DISEASE (ES=0.60000, NP=0.0173), APICAL_JUNCTION(ES=0.5524, NP=0.0404), REGULATED_NECROSIS (E=0.5397, NP=0.0376);
ALZHEIMERS_DISEASE (ES=0.60000, NP=0.0173), APICAL_JUNCTION(ES=0.5524, NP=0.0404), REGULATED_NECROSIS (E=0.5397, NP=0.0376);  PROTEASOME (ES=0.3176, NP=0.0347), TGF_BETA_SIGNALING (ES=0.3726, NP=0.0479), APOPTOSIS (ES=0.3806, NP=0.0382);
PROTEASOME (ES=0.3176, NP=0.0347), TGF_BETA_SIGNALING (ES=0.3726, NP=0.0479), APOPTOSIS (ES=0.3806, NP=0.0382);  PATHOGENIC ESCHERICHIA COIL INFECTION (ES=0.3847, NP=0.0285), MYC TARGETS VI (ES=0.3874, NP=0.0486), PROGRAMMED CELL DEATHS (ES=0.3927, NP=0.0377)
PATHOGENIC ESCHERICHIA COIL INFECTION (ES=0.3847, NP=0.0285), MYC TARGETS VI (ES=0.3874, NP=0.0486), PROGRAMMED CELL DEATHS (ES=0.3927, NP=0.0377)
To further elucidate the biological functions assumed by the OS_ PAN_DEGs, we performed GSEA analysis on the OS_PAN_DEGs using the KEGG, hallmark, and reactome datasets. The results suggested that the OS_PAN_DEGs were primarily enriched in apoptosis (reactome and KEGG), mammalian Target Of Rapamycin Complex 1 (mTORC1) signaling pathway (hallmark), focal adhesion (KEGG), and Vascular Endothelial Growth Factor (VEGF) signaling pathway (reactome) (NES>1, p-value <0.05, FDR<0.25), (Figures 2G-2I). Overall, our study identified 105 PANoptosis-related differentially expressed genes (OS_ PAN_DEGs) in OS and revealed their enrichment in various biological functions and signaling pathways, such as the mTORC1 signaling pathway, apoptosis, and VEGF signaling pathway, indicating their potential involvement in tumor growth, metastasis and drug resistance (Figures 4A-4E).
Identification of three OS subtypes with different prognoses based on the expression profile of OS_PAN_DEGs
Previous studies have indicated that PANoptosis may influence tumor mutations and immune infiltration. To assess the immune landscape across subgroups of OS_PAN_DEGs, we utilized several immune algorithms, including cibersort, estimate, and xcell to analyze the immune profiles of the three clusters. A waterfall plot in Figure 5A depicts the distribution of 22 immune cell types within the training set of GSE21257. Subsequently, we evaluated the immune-score, stromal-score and microenvironment score across the subgroups of OS_PAN_DEGs (Figure 5B and 5C). Our findings revealed significant differences in both immune-score and stromal-score between cluster_1 and the other two groups (cluster_1 vs. cluster_2, P=0.006; cluster_1 vs. cluster_3, P=0.043 for immune-score; cluster_1 vs. cluster_2, P=0.0066; cluster_1 vs. cluster_3, P=0.016 for stromal-score). In the assessment of microenvironment score, no statistical difference was observed between cluster_1 and cluster_3, whereas a significant difference was present between cluster_1 and cluster_2 (P=0.0023), (Figure 5D). Furthermore, we evaluated the gene expression of immune checkpoints in the subgroups of OS_PAN_DEGs, including (CD276), Cluster of Differentiation 86 (CD86), Tumor Necrosis Factor Ligand Superfamily Member 14 (TNFSF14), Cluster of Differentiation 40 (CD40), Cluster of Differentiation 48 (CD48), Hepatitis A Virus Cellular Receptor 2 (HAVCR2), Leukocyte Associated Immunoglobulin Like Receptor 1 (LAIR1), Programmed Cell Death 1 Ligand 2 (PDCD1LG2) and Tumor Necrosis Factor Ligand Superfamily Member 4 TNFSF4. These immune checkpoints were upregulated in cluster_2 and down regulated in cluster_1 and cluster_3 (all P<0.05), (Figure 5E). Subsequently, we presented the somatic mutation signatures across the subgroups of OS_ PAN_DEGs. The top 19 genes with the highest mutation frequencies across the three clusters were Tumor Protein 53 (TP53), Mucin 16, Cell Surface Associated (MUC16), Alpha Thalassemia/Mental Retardation Syndrome X-Linked (ATRX), Retinoblastoma 1 (RB1), Contactin Associated Protein-Like 5 (CNTNAP5), Titin (TTN), CUB and Sushi Multiple Domains 3 (CSMD3), Piccolo Presynaptic Cytomatrix Protein (PCLO), Alström Syndrome Protein 1 (ALMS1), Dystrophin (DMD), Dynein Axonemal Heavy Chain 3 (DNAH3), HECT Domain E3 Ubiquitin Protein Ligase 4 (HECTD4), Laminin Subunit Alpha 2 (LAMA2), Ryanodine Receptor 2 (RYR2), Calcium Voltage-Gated Channel Subunit Alpha1 B (CACNA1B), Chromodomain Helicase DNA Binding Protein 1-Like (CHD1L), CUB and Sushi Multiple Domains 2 (CSMD2), CXXC Finger Protein 1 (CXXC1), Dynein Axonemal Heavy Chain 9 (DNAH9), and Dynein Axonemal Intermediate Chain 4 (DNAI4). Among these, Tumor Protein 53 (TP53) had the highest mutation frequency (21%), while the mutation frequencies of the other nine genes ranged from 3%-11% (Figure 5F). The most common mutations involved cytosine, indicating cytosine instability, potentially due to the ease of oxidation of its amino group. Tumor Mutation Burden (TMB), an important indicator of the number of mutations in cancer, is also a novel biomarker for assessing the response to PD-1 antibody therapy. We compared the TMB across the three clusters and found that cluster_2 had a higher TMB than cluster_1 and cluster_3 (cluster_2 vs. cluster_1, P=0.00017; cluster_2 vs. cluster_3, P=0.0019), (Figure 5G). In summary, our findings reveal significant differences in the expression of immune checkpoints and mutation signatures across the three subgroups, which may have important implications for cancer immunotherapy.
Figure 4: Identification of three hepatocellular carcinoma subtypes with different prognoses using OS_PAN_DEG expression profiles. (A, B): Evaluation of the average silhouette width and area under the CDF curve within clusters when k=2 to 10; ©: Classification of the training cohort into three osteosarcoma subtypes using consensus clustering; (D): Heat map showing the expression of OS_PAN_DEGs in different osteosarcoma subtypes; (E): Kaplan-Meier survival analysis among the three OS_PAN_DEGs subtypes.
Constructing a risk assessment model
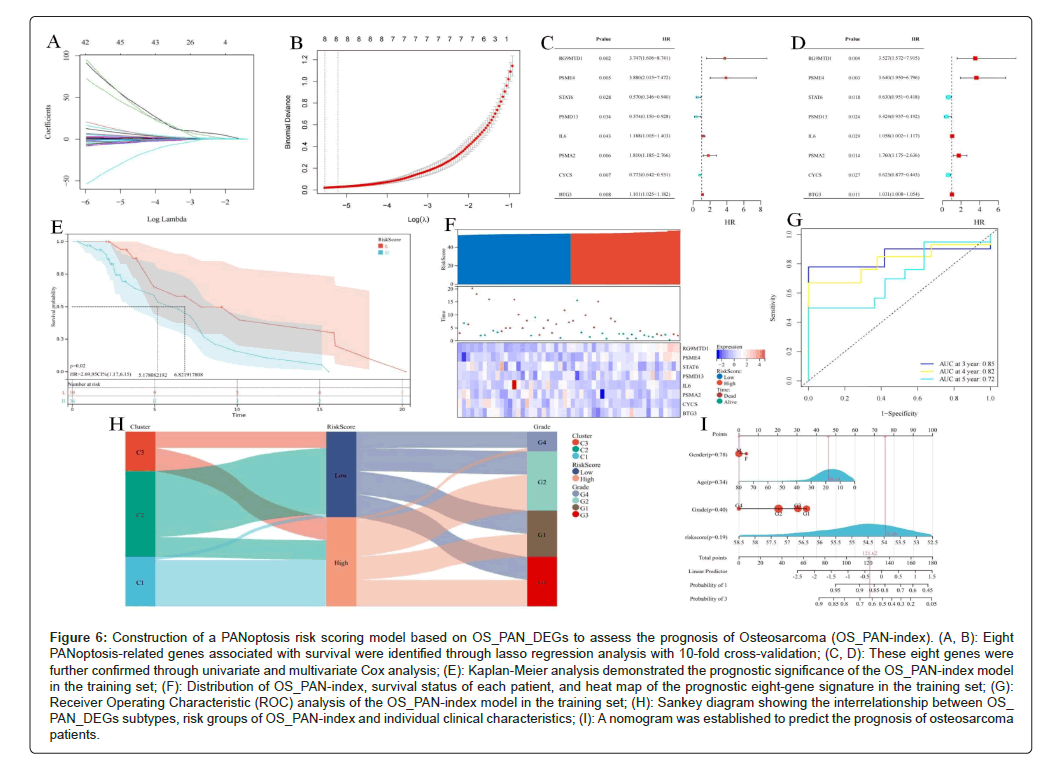
To further assess the impact of OS_PAN_DEGs on survival prognosis, we utilized Least Absolute Shrinkage and Selection Operator (LASSO), univariate, and multivariate regression to screen out eight gene signatures closely related to prognosis. Based on these signatures, we ultimately constructed a PANoptosis risk index model (OS_PAN- index) for OS (Figures 6A-6D). The formula for the OS_PAN-index is as follows: HPAN-inde = 0.188BTG3-0.288CYCS+0.246DYNLL1-0.235IL 6+0.898PSMA2+0.587PSMD13+0.673PSME4+0.321STAT6. Using the OS_PAN-index score, we classified 53 patients with complete survival information in the training set into a high OS_PAN-index group and a low OS_PAN-index group (34 in the high group vs. 19 in the low group). Kaplan-Meier analysis revealed that the prognosis of the low- risk group (Median Survival Time, MST=6.82 years) in the training set was significantly better than that of the high-risk group (MST=5.18 years) (P=0.02), (Figure 6E). We investigated the relationship between patient prognosis, gene expression, and the OS_PAN-index, observing a significant decrease in survival rates with increasing OS_PAN-index. As expected STAT6, IL6, PSMA2, CYCS emerged as protective factors, with their expressions downregulated as the OS_PAN-index GDI, RG9MTD1, PSME4, PSMD13, BTG3 were identified as risk factors (Figure 6F). Additionally, we evaluated the Area Under the Curve (AUC) of the OS_PAN-index as a predictive model, demonstrating its high accuracy in predicting 3-year, 4-year, and 5-year survival (Figure 6G).
Figure 5: Unique immune characteristics and mutation landscapes in the OS_PAN_DEGs subgroups. A: Waterfall plot of the distribution of 22 immune cell types in the training set; B-D: Immune scores, stromal scores, and microenvironment scores for the three HPAN_DEGs subgroups; E: Expression of immune check points in different HPAN_DEGs subtypes; F: Mutation landscape of OS_PAN_DEGs alterations between high-risk and low-risk osteosarcoma patients in the training set; G: Tumor mutational burden indifferent OS_PAN_DEGs subtypes.
Note: 
Sankey diagrams were employed to visualize the relationship between the OS_PAN-index risk groups and individual clinical characteristics, revealing that cluster_1 was mainly concentrated in the high OPANindex group, while cluster_2 was primarily found in the low OS_PAN-index group (Figure 6H). Based on the results of cox regression analysis, we constructed a nomograph, which identified the OS_PAN-index as an independent risk factor (Figure 6I). In summary, the PANoptosis risk scoring model (OPAN-index) based on OS_PAN_DEGs, constructed through LASSO regression, univariate and multivariate regression analysis, accurately predicts the survival prognosis of OS patients and has the potential to become a potential independent risk factor for clinical decision-making.
Figure 6: Construction of a PANoptosis risk scoring model based on OS_PAN_DEGs to assess the prognosis of Osteosarcoma (OS_PAN-index). (A, B): Eight PANoptosis-related genes associated with survival were identified through lasso regression analysis with 10-fold cross-validation; (C, D): These eight genes were further confirmed through univariate and multivariate Cox analysis; (E): Kaplan-Meier analysis demonstrated the prognostic significance of the OS_PAN-index model in the training set; (F): Distribution of OS_PAN-index, survival status of each patient, and heat map of the prognostic eight-gene signature in the training set; (G): Receiver Operating Characteristic (ROC) analysis of the OS_PAN-index model in the training set; (H): Sankey diagram showing the interrelationship between OS_ PAN_DEGs subtypes, risk groups of OS_PAN-index and individual clinical characteristics; (I): A nomogram was established to predict the prognosis of osteosarcoma patients.
Validating the effectiveness of the OS_PAN-index as a predictive factor for the prognosis of OS patients
To test the reproducibility of the OS_PAN-index model as a predictive tool, we validated the model in the TARGET-OS cohort (validation set). Using Kaplan-Meier analysis, we observed a significant decrease in patient survival rates with increasing OS_PAN-index (Figure 7A). In the validation set, the prognosis of the low OS_PAN- index group was significantly better than that of the high OS_PAN- index group (MST=1.79 years vs MST=10.89 years, P<0.001) (Figure 7B). In summary, the OS_PAN-index model demonstrated significant predictive value for patient survival rates in the validation set, with higher OS_PAN-index scores correlating with poorer prognoses.
Figure 7: Group validation of OS_PAN-index as a prognostic predictor in Osteosarcoma patients. (A): Distribution of OS_PAN-index, survival status of each patient, and heat map of the prognostic eight-gene signature in the validation set (target-OS, n=95); (B): Kaplan-Meier analysis demonstrated the prognostic significance of the OS_PAN-index model in the validation set; (C): Boxplots showing significant differences in immune cells between different OS_PAN-index groups; (D): Correlation between OS_PAN-index and immune cell infiltration in osteosarcoma assessed using cibersort; (E): Immune scores, stromal scores, and estimate scores among different OS_PAN-index groups. *p:<0.05; **: p<0.0; ***: p<0.001; ****: p<0.0001.
Note: 
Immune cell profiles and molecular pathways associated with the OS_PAN-index in OS patients
To further explore the immune characteristics of different OS_PAN- index groups, we employed five distinct immune algorithms, including cibersort, estimate, tide, timer, and xcell, to assess the relationship between the OS_PAN-index and the immune microenvironment. The results in Figures 7C and 7D indicate a significant correlation between the OS_PAN-index and the expression of Plasma cells, T cells gamma delta, and NK cells resting, with all P-values less than 0.05. Additionally, the stromal score, immune score, and estimate score were significantly higher in the high OS_PAN-index group compared to the low OS_ PAN-index group (all P<0.05), (Figure 7E). Notably, the Microsatellite Instability (MSI) score, an important indicator reflecting tumor genomic stability, is closely associated with the effectiveness of immune checkpoints (56). Our analysis revealed a significantly higher MSI score in the high OS_PANindex group compared to the low OS_PAN-index group, with similar results observed for other scores such as the Tumor Immune Dysfunction and Exclusion (TIDE) score, Interferon-Gamma (IFNG) score, Cluster of Differentiation 86 CD8 score, dysfunction score, exclusion score, Myeloid-Derived Suppressor Cells (MDSC) score, and Tumor-Associated Macrophage (TAM) score (all P<0.05), (Figures 8A-8H). This observation suggests that the high OS_PAN- index group may be more likely to benefit from immunotherapy compared to the low OS_PAN-index group.
Figure 8: Immune cell landscape and molecular pathways associated with OS_PAN-index in osteosarcoma patients. A-H: Tide score, IFNG score, CD8 score, dysfunction score, exclusion score, MDSC score, and TAM score in low and high OS_PAN-index groups; I: Enriched pathways for genes with specific expression in the high and low OS_PAN-index groups.
To further elucidate the molecular mechanisms underlying different OS_PAN-index groups, we conducted Gene Set Variation Analysis (GSVA) enrichment analysis. The results indicated significant enrichment in signaling pathways such as spliceosome, ubiquitin- mediated proteolysis, mismatch repair, and steroid biosynthesis in the high OS_PAN-index group. In contrast, the low OS_PAN- index group exhibited significant enrichment in signaling pathways related to glycosaminoglycan degradation, glycerolipid metabolism, alanine, aspartate, and glutamate metabolism, and glycosphingolipid biosynthesis (Figure 8I). These findings suggest that patients with higher OS_PAN-index scores may be more sensitive to immune checkpoint therapy, while those with lower OS_PAN-index scores may be more suitable for targeted drug therapy.
Drug screening
To identify potential drugs for the treatment of OS, we conducted a drug target enrichment analysis using the cellminer and DsigDB databases, focusing on eight core genes BTG3, CYCS, DYNLL1, IL6, PSMA2, PSMD13, PSME4, and STAT6 as genetic targets relevant to risk model construction. The results revealed a significant positive correlation between the expression levels of these eight core genes and the drugs pazopanib, chelerythrine, staurosporine, hydroxyurea and sunitinib (Table 2). These drugs provide important reference points for the development of OS treatment strategies.
Discussion
OS, a highly malignant tumor often occurring in children and adolescents, exhibits remarkable infiltrative and metastatic capabilities [42]. Currently, traditional treatment methods for OS have demonstrated limitations, such as the limited killing effect of chemotherapy on tumor cells, which can easily lead to the development of drug resistance [43]. Therefore, the search for novel therapeutic approaches and targets holds significant importance for the treatment of OS.
Recently, the concept of PANoptosis has garnered widespread attention in the biomedical field. As an inflammatory form of programmed cell death mediated by the PANoptosome complex, PANoptosis encompasses all the key features of pyroptosis, necroptosis, and apoptosis, making it a important breakthrough in the study of cell death mechanisms [44]. In the development of OS, a malignant bone tumor, PANoptosis plays a major role. OS, originating from the mesenchymal cell lineage, is highly malignant with a poor prognosis, often leading to lung metastasis within a few months, posing a severe threat to patients’ lives and health [45]. The characteristics of PANoptosis make it significan in the immune response and tumor progression of OS. Although previous studies have gained some understanding of the molecular mechanisms of necroptosis [46,47], pyroptosis, and apoptosis in OS they often focus on only one or two forms of cell death, failing to comprehensively assess their roles in OS Our study is the first attempt to evaluate the characteristics of PANoptosis in OS.
In this study, we examine into the characteristics and roles of PANoptosis in OS for the first time. Through the normalization of GEO series combined gene expression data, we screened out a gene set highly related to OS. Subsequently, using WGCNA, we identified the most relevant module for OS. By integrating information from GEO, WGCNA, and the PANoptosis gene set, we identified 105 OS_ PAN_DEGs related to PANoptosis. These genes not only encompassed the core processes of PANoptosis, such as pyroptosis, apoptosis, and necroptosis, but were also closely associated with the occurrence and development of OS. The results of GO, KEGG, and GSEA enrichment analyses revealed that OS_PAN_DEGs were involved in various pathways, including cell metabolic regulation, hematopoietic stem cell differentiation, interleukin-1-mediated signaling, RNA transcriptional regulation, apoptosis, necroptosis, proteasome, hippo signaling, and neurodegenerative diseases. Notably, we found that OS_PAN_DEGs were particularly enriched in the Mammalian Target of Rapamycin Complex 1 (mTORC1) signaling pathway and VEGF signaling pathway. mTORC1, as a key regulator of cell growth and metabolism, its abnormal activation is often closely associated with tumorigenesis and development [48]. Therefore, we speculate that OS_PAN_DEGs may affect the proliferation and metabolism of OS cells by regulating the mTORC1 signaling pathway, thereby promoting tumor growth and metastasis. In addition, we also observed a significant enrichment of OS_PAN_DEGs in the apoptotic process. Apoptosis is an important form of programmed cell death, and its abnormality often leads to the unlimited proliferation and malignant transformation of tumor cells [49]. By further studying the specific mechanisms of these genes in apoptosis, we hope to provide new targets for the treatment of OS.
Utilizing consensus clustering analysis, we successfully classified the OS cohort into three subtypes with distinct prognoses based on the expression profile information of OS_PAN_DEGs. The expression of PSMD14 and PPT1 genes was upregulated in cluster_1, while TIMP1 and GADD45A were down regulated in cluster_2 and cluster_3 relative to cluster_1. These differences in gene expression may directly affect the apoptotic process of OS cells, resulting in prognostic variations among the different subtypes. cluster_2 exhibited higher immune and stromal scores, as well as a higher TMB. These characteristics may render cluster_2 more sensitive to immunotherapy, potentially explaining its relatively better overall survival rate [50]. Additionally, differences in the expression of immune checkpoints among the different subtypes suggest the possibility of developing personalized immunotherapy strategies tailored to each subtype of OS. Notably, our study also identified several genes with high-frequency mutations, such as TP53. Mutations in these genes may influence the biological behaviors of OS cells, including proliferation, apoptosis, invasion, and metastasis [51]. Further investigation of these mutated genes can not only reveal the pathogenesis of OS but also provide clues for the development of novel targeted therapies.
Through LASSO regression, univariate, and multivariate regression analysis, we identified eight gene signatures closely related to prognosis and subsequently constructed the OS_PAN-index scoring system. This scoring system demonstrated excellent predictive performance in the training set, with the prognosis of the high-risk group being significantly worse than that of the low-risk group. In the validation set, we further validated the effectiveness of the OS_PAN-index, observing a significant decrease in patient survival rates with increasing OS_PAN- index. We delved into the relationship between the OS_PAN-index and the immune microenvironment, finding significant correlations between high OS_PAN-index groups and the expression of various immune cell types. Moreover, the immune scores, stromal scores, and microenvironment scores were higher in these groups compared to the low OS_PANindex groups. Additionally, we observed a significant increase in MSI scores and other scores related to immunotherapy effectiveness in the high OS_PAN-index groups. These results suggest that OS patients with high OS_PAN-index may be more likely to benefit from immunotherapy. Through GSVA enrichment analysis, we further revealed the molecular mechanisms underlying different OS_PAN-index groups. The high OS_PAN-index groups exhibited significant enrichment in signaling pathways such as the spliceosome and ubiquitin-mediated protein hydrolysis, which are typically associated with tumor immune responses and cell death processes[52]. In contrast, the low OS_PAN-index groups were enriched in some metabolic-related pathways. These results not only aid in understanding the biological differences between different OS_PAN-index groups but also provide a theoretical basis for developing personalized treatment strategies for OS patients based on their OS_PAN-index.
Finally, utilizing the cellminer and DsigDB databases, we conducted a drug target enrichment analysis for the eight core genes closely related to the construction of the OS risk model. The analysis revealed significant positive correlations between the expression levels of these core genes and drugs such as pazopanib, chelerythrine, staurosporine, hydroxyurea, and sunitinib. Pazopanib, a multi-target tyrosine kinase inhibitor, has demonstrated antitumor activity in various tumors by inhibiting tumor angiogenesis and tumor cell proliferation [53]. Chelerythrine, a natural product extracted from plants, exhibits a wide range of biological activities, including antitumor effects [54]. Staurosporine, a broad-spectrum protein kinase inhibitor, can induce apoptosis and inhibit multiple tumors [55]. Hydroxyurea, a commonly used cytotoxic drug, exerts its therapeutic effects by inhibiting DNA synthesis [56]. Sunitinib, another multitarget tyrosine kinase inhibitor, is widely used in the treatment of renal cell carcinoma and has shown efficacy in other solid tumors as well [57]. The discovery of these drugs provides important references for developing therapeutic strategies for OS. Considering the complexity and heterogeneity of OS, a single treatment approach often fails to achieve satisfactory results. Therefore, combining the characteristics of these potential drugs, we can consider developing combined treatment regimens in anticipation of achieving better therapeutic outcomes.
When discussing this study, we must acknowledge some existing limitations. Although we have made every effort to analyze the immune landscape of OS through bulk RNA sequencing data, the lack of single-cell sequencing data and spatial transcriptomic data remains a significant shortcoming. The immune microenvironment is a highly complex and intricate microsystem, where cellular differences and interactions are necessary for understanding disease progression and developing therapeutic strategies. Therefore, relying solely on macroscopic bulk data for analysis may result in the loss of major cellular level information. Furthermore, to more accurately validate the predictive value of our OS_PAN-index model in immunotherapy for OS, we require additional immunotherapy-related data with larger sample sizes. Such data would provide more direct and compelling evidence to support our model.
Conclusion
In summary, we conducted a thorough exploration of the OS_ PAN_DEGs related to the PANoptosis process in OS, successfully identifying three subgroups with unique prognostic features, mutation patterns, and immune infiltration characteristics. Notably, these subgroups also exhibited significant differences in the expression of immune checkpoints, deepening our understanding of the biological characteristics of OS.Furthermore, we constructed the OS_PAN index, an innovative indicator that is not only closely related to patient survival prognosis but also effectively predicts patient responses to smallmolecule targeted drugs and immunotherapy. We anticipate that this model will assist doctors in more accurately identifying patients suitable for immunotherapy or targeted therapy, providing new strategies for personalized treatment of OS. This study not only opens a new path for precision medicine in OS but may also provide new insights for revealing the PANoptosis mechanism in OS.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent to participate declaration
Every participant has voluntarily agreed to participate in this study
Consent for publication
Not applicable.
Data availability declaration
Not applicable.
Ethics approval declaration
Not applicable.
Funding
No funding was received related to the writing and publication of this manuscript.
Authors’ contributions
Qian Zhang, Qishuai Zhuang designed the study. Song Zhou, Jing Zhou, Lianxiang Li, Bo Song, Yuelei Cheng, Wei Xie, Yunlai Zhao, Feng Yang performed all the experiments and analyzed the data. Song Zhou and Jing Zhou wrote the first draft of the manuscript. Qian Zhang, Qishuai Zhuang reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Declaration of competing interest
All authors disclosed no relevant relationships.
Acknowledgements
Not applicable.
References
- Isakoff MS, Bielack SS, Meltzer P, Gorlick R (2005) Osteosarcoma: Current treatment and a collaborative pathway to success. J Clin Oncol 33:3029-3035.
[Crossref] [Google Scholar] [PubMed]
- Gang W, Hao H, Yong H, Ruibing F, Chaowen L, et al. (2022) Therapeutic potential of triptolide intreating bone-related disorders. Front Pharmacol 13:905576.
[Crossref] [Google Scholar] [PubMed]
- Akkawi R, Hidmi O, Haj-Yahia A, Monin J, Diment J, et al. (2024) WWOX promotes osteosarcoma development via upregulation of Myc. Cell Death Dis 15:13.
[Crossref] [Google Scholar] [PubMed]
- Heng M, Gupta A, Chung PW, Healey JH, Vaynrub M, et al. (2020) The role of chemotherapy and radiotherapy in localized extra skeletal osteosarcoma. Eur J Cancer 125:130-141.
[Crossref] [Google Scholar] [PubMed]
- Harris CJ, Helenowski I, Murphy AJ, Mansfield SA, La Quaglia MP, et al. (2022) Implications of tumor characteristics and treatment modality on local recurrence and functional outcomes in children with chest wall sarcoma: A pediatric surgical oncology research collaborative study. Ann Surg 276:e969-e975.
[Crossref] [Google Scholar] [PubMed]
- Strauss SJ, Frezza AM, Abecassis N, Bajpai J, Bauer S, et al. (2021) Bone sarcomas: ESMO-EURACAN-GENTURIS-ERN Paed can clinical practice guideline for diagnosis, treatment and follow-up. Ann Oncol 32:1520-1536.
[Crossref] [Google Scholar] [PubMed]
- Xiao X, Wang W, Li Y, Yang D, Li X, et al. (2018) HSP90AA1-mediated autophagy promotes drug resistance in osteosarcoma. J Exp Clin Cancer Res 37:201.
[Crossref] [Google Scholar] [PubMed]
- Qi Z, Zhu L, Wang K, Wang N (2023) PANoptosis: Emerging mechanisms and disease implications. Life Sci 333:122158.
[Crossref] [Google Scholar] [PubMed]
- Karki R, Sharma BR, Tuladhar S, Williams EP, Zalduondo L, et al. (2021) Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell 184:149-168.e17.
[Crossref] [Google Scholar] [PubMed]
- Jiang M, Qi L, Li L, Wu Y, Song D, et al. (2021) Caspase-8: A key protein of cross-talk signal way in "PANoptosis" in cancer. Int J Cancer 149:1408-1420.
[Crossref] [Google Scholar] [PubMed]
- Cai Y, Chen X, Lu T, Fang X, Ding M, et al. (2023) Activation of STING by SAMHD1 deficiency promotes PANoptosis and enhances efficacy of PD-L1 blockade in diffuse large B-cell lymphoma. Int J Biol Sci 19:4627-4643.
[Crossref] [Google Scholar] [PubMed]
- Ishida Y, Agata Y, Shibahara K, Honjo T (1992) Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J 11:3887-3895.
[Crossref] [Google Scholar] [PubMed]
- Zhang M, Huang Y, Zou J, Yang Y, Yao Y, et al. (2023) Advanced oxidation nano processing boosts immunogenicity of whole tumor cells. Adv Sci10:e2302250.
- Li J, Yang R, Dong Y, Chen M, Wang Y, et al. (2019) Knockdown of FOXO3a induces epithelial-mesenchymal transition and promotes metastasis of pancreatic ductal adenocarcinoma by activation of the β-catenin/TCF4 pathway through SPRY2. J Exp Clin Cancer Res 38:38.
[Crossref] [Google Scholar] [PubMed]
- Xu J, Shen W, Pei B, Wang X, Sun D, et al. (2018) Xiao Tan He Wei decoction reverses MNNG-induced precancerous lesions of gastric carcinoma in vivo and vitro: Regulation of apoptosis through NF-κB pathway. Biomed Pharmacother 108:95-102.
[Crossref] [Google Scholar] [PubMed]
- Morad G, Helmink BA, Sharma P, Wargo JA (2021) Hallmarks of response, resistance and toxicity to immune checkpoint blockade. Cell 184:5309-5337.
[Crossref] [Google Scholar] [PubMed]
- Blomberg OS, Spagnuolo L, Garner H, Voorwerk L, Isaeva OI, et al. (2023) IL-5-producing CD4+ Tcells and eosinophils cooperate to enhance response to immune checkpoint blockade in breast cancer. Cancer cell 41:106-123.e10.
[Crossref] [Google Scholar] [PubMed]
- Tichet M, Wullschleger S, Chryplewicz A, Fournier N, Marcone R, et al. (2023) Bispecific PD1-IL2v and anti-PD-L1 break tumor immunity resistance by enhancing stem-like tumor-reactive CD8+ Tcells and reprogramming macrophages. Immunity 56:162-179.e6.
[Crossref] [Google Scholar] [PubMed]
- Zhou Y, Yang D, Yang Q, Lv X, Huang W, et al. (2020) Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat Commun 11:6322.
[Crossref] [Google Scholar] [PubMed]
- Lv Y, Wu L, Jian H, Zhang C, Lou Y, et al. (2022) Identification and characterization of aging/senescence induced genes in osteosarcoma and predicting clinical prognosis. Front Immunol. 13:997765.
[Crossref] [Google Scholar] [PubMed]
- Yang JZ, Ma SR, Rong XL, Zhu MJ, Ji QY, et al. (2016) Characterization of multidrug‑resistant osteosarcoma sublines and the molecular mechanisms of resistance. Mol Med Rep 14:3269-3276.
[Crossref] [Google Scholar] [PubMed]
- Liu W, Hu H, Shao Z, Lv X, Zhang Z, et al. (2023) Characterizing the tumor microenvironment at the single-cell level reveals a novel immune evasion mechanism in osteosarcoma. Bone Res 11:4.
[Crossref] [Google Scholar] [PubMed]
- Wang Z, Li B, Li S, Lin W, Wang Z, et al. (2022) Metabolic control of CD47 expression through LAT2-mediated amino acid uptake promotes tumor immune evasion. Nat Commun 13:6308.
[Crossref] [Google Scholar] [PubMed]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, et al. (2013) NCBI GEO: Archive for functional genomics datasets-update. Nucleic Acids Res 41:D991-D995.
[Crossref] [Google Scholar] [PubMed]
- Paoloni M, Davis S, Lana S, Withrow S, Sangiorgi L, et al. (2009) Canine tumor cross-species genomics uncovers targets linked to osteosarcoma progression. BMC Genomics 10:625.
[Crossref] [Google Scholar] [PubMed]
- Buddingh EP, Kuijjer ML, Duim RA, Bürger H, Agelopoulos K, et al. (2011) Tumor-infiltrating macrophages are associated with metastasis suppression in high-grade osteosarcoma: A rationale for treatment with macrophage activating agents. Clin Cancer Res 17:2110-2119.
[Crossref] [Google Scholar] [PubMed]
- Fritsche-Guenther R, Noske A, Ungethüm U, Kuban RJ, Schlag PM, et al. (2010) De novo expression of EphA2 in osteosarcoma modulates activation of the mitogenic signaling pathway. Histopathology 57:836-850.
[Crossref] [Google Scholar] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. (2005) Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545-15550.
[Crossref] [Google Scholar] [PubMed]
- Langfelder P, Horvath S (2008) WGCNA: An R package for weighted correlation network analysis. BMC Bio informatics 9:559.
[Crossref] [Google Scholar] [PubMed]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, et al. (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47.
[Crossref] [Google Scholar] [PubMed]
- Yu W, Ma Y, Hou W, Wang F, Cheng W, et al. (2021) Identification of immune-related lncRNA prognostic signature and molecular subtypes for glioblastoma. Front Immunol 12:706936.
[Crossref] [Google Scholar] [PubMed]
- Hein DM, Deng W, Bleile M, Kazmi SA, Rhead B, et al. (2022) Racial and ethnic differences in genomic profiling of early onset colorectal cancer. J Natl Cancer Inst 114:775-778.
[Crossref] [Google Scholar] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. (2005) Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc NatlAcad Sci USA 102:15545-15550.
[Crossref] [Google Scholar] [PubMed]
- Niknafs N, Balan A, Cherry C, Hummelink K, Monkhorst K, et al. (2023) Persistent mutation burden drives sustained anti-tumor immune responses. Nat Med 29:440-449.
[Crossref] [Google Scholar] [PubMed]
- Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP (2018) Maftools: Efficient and comprehensiveanalysis of somatic variants in cancer. Genome Res 28:1747-1756.
[Crossref] [Google Scholar] [PubMed]
- Wang T, Dai L, Shen S, Yang Y, Yang M, et al. (2022) Comprehensive molecular analyses of a macrophage-related gene signature with regard to prognosis, immune features and biomarkers for immunotherapy in hepatocellular carcinoma based on wgcna and the lasso algorithm. Front Immunol 13:843408.
[Crossref] [Google Scholar] [PubMed]
- Chen X, Lan H, He D, Xu R, Zhang Y, et al. (2021) Multi-omics profiling identifies risk hypoxia-related signatures for ovarian cancer prognosis . Front Immunol 12:645839.
[Crossref] [Google Scholar] [PubMed]
- Charras G, Lenz M (2022) A biochemical timer phases condensates in and out in cells. Nature 609:469-470.
[Crossref] [Google Scholar] [PubMed]
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, et al. (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12:453-457.
[Crossref] [Google Scholar] [PubMed]
- Yoo M, Shin J, Kim J, Ryall KA, Lee K, et al. (2015) DSigDB: Drug signatures database for gene set analysis. Bioinformatics 31:3069-3071.
[Crossref] [Google Scholar] [PubMed]
- Shankavaram UT, Varma S, Kane D, Sunshine M, Chary KK, et al. (2009) Cellminer: A relational database and query tool for the NCI-60 cancer cell lines. BMC Genomics 10:277.
[Crossref] [Google Scholar] [PubMed]
- Ligon JA, Choi W, Cojocaru G, Fu W, Hsiue EH, et al. (2021) Pathways of immune exclusion in metastatic osteosarcoma are associated with inferior patient outcomes. J Immunother Cancer 9:e001772.
[Crossref] [Google Scholar] [PubMed]
- Lu B, Zou C, Yang M, He Y, He J, et al. (2021) Pharmacological inhibition of core regulatory circuitry liquid‒liquid phase separation suppresses metastasis and chemo resistance in osteosarcoma. Adv Sci 8:e2101895.
[Crossref] [Google Scholar] [PubMed]
- Sundaram B, Pandian N, Mall R, Wang Y, Sarkar R, et al. (2023) NLRP12-PANoptosome activates PANoptosis and pathology in response to heme and PAMPs. Cell 186:2783-2801.e20.
[Crossref] [Google Scholar] [PubMed]
- Liu W, Zhao Y, Wang G, Feng S, Ge X, et al. (2022) TRIM22 inhibits osteosarcoma progression through destabilizing NRF2 and thus activation of ROS/AMPK/mTOR/autophagy signaling. Redox Biol 53:102344.
[Crossref] [Google Scholar] [PubMed]
- Jin J, Yuan P, Yu W, Lin J, Xu A, et al. (2022) Mitochondria-targeting polymer micelle of dichloroacetate induced pyroptosis to enhance osteosarcoma immunotherapy. ACS Nano 16:10327-10340.
[Crossref] [Google Scholar] [PubMed]
- Li S, Zhang T, Xu W, Ding J, Yin F, et al. (2020) Sarcoma-targeting peptide-decorated polypeptidenanogel intracellularly delivers shikonin for upregulated osteosarcoma necroptosis and diminished pulmonary metastasis. Theranostics 8:1361-1375.
[Crossref] [Google Scholar] [PubMed]
- Zoncu R, Efeyan A, Sabatini DM (2011) mTOR: From growth signal integration to cancer, diabetes and aging. Nat Rev Mol Cell Biol 12:21-35.
[Crossref] [Google Scholar] [PubMed]
- Cacchione S, Biroccio A, Rizzo A (2019) Emerging roles of telomeric chromatin alterations in cancer. J Exp Clin Cancer Res 38:21.
[Crossref] [Google Scholar] [PubMed]
- Litchfield K, Reading JL, Puttick C, Thakkar K, Abbosh C, et al. (2021) Meta-analysis of tumor-and T-cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell 184:596-614.e14.
[Crossref] [Google Scholar] [PubMed]
- Gourisankar S, Krokhotin A, Ji W, Liu X, Chang CY, et al. (2023) Rewiring cancer drivers to activate apoptosis. Nature 620:417-425.
[Crossref] [Google Scholar] [PubMed]
- Xu S, Feng Z, Zhang M, Wu Y, Sang Y, et al. (2011) hSSB1 binds and protects p21 from ubiquitin-mediated degradation and positively correlates with p21 in human hepatocellular carcinomas. Oncogene 30:2219-2229.
[Crossref] [Google Scholar] [PubMed]
- Sherman EJ, Harris J, Bible KC, Xia P, Ghossein RA, et al. (2023) Radiotherapy and paclitaxel pluspazopanib or placebo in anaplastic thyroid cancer (NRG/RTOG 0912): A randomized, double-blind, placebo-controlled, multicentre, phase 2 trial. Lancet Oncol 24:175-186.
[Crossref] [Google Scholar] [PubMed]
- Wang P, Zheng SY, Jiang RL, Wu HD, Li YA, et al. (2023) Necroptosis signaling and mitochondrial dysfunction cross-talking facilitate cell death mediated by chelerythrine in glioma. Free Radic Biol Med 202:76-96.
[Crossref] [Google Scholar] [PubMed]
- Short NJ, Rytting ME, Cortes JE (2018) Acute myeloid leukemia. Lancet 392:593-606.
[Crossref] [Google Scholar] [PubMed]
- Andrs M, Stoy H, Boleslavska B, Chappidi N, Kanagaraj R, et al. (2023) Excessive reactive oxygen species induce transcription-dependent replication stress. Nat Commun 14:1791.
[Crossref] [Google Scholar] [PubMed]
- Motzer RJ, Powles T, Burotto M, Escudier B, Bourlon MT, et al. (2022) Nivolumab plus cabozantinib versus sunitinib in first-line treatment for advanced renal cell carcinoma (CheckMate 9ER): Long-term follow-up results from an open-label, randomized, phase 3 trial. Lancet Oncol 23:e319.
[Crossref] [Google Scholar] [PubMed]
Citation: Zhou S, Zhou J, Li L, Song B, Cheng Y, et al. (2024) Developing a PANoptosis Signature: Identification of Unique Immunotherapeutic Candidates for Osteosarcoma. J Infect Dis Ther 12:598. DOI: 10.4172/2332-0877.1000598
Copyright: © 2024 Zhou S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 895
- [From(publication date): 0-2024 - Apr 26, 2025]
- Breakdown by view type
- HTML page views: 696
- PDF downloads: 199