Determining the Level of Genotypic Variability of different Upland Rice Genotypes using Cluster Analysis
Received: 28-Sep-2022 / Manuscript No. rroa-22-71450 / Editor assigned: 03-Oct-2022 / PreQC No. rroa-22-71450 / Reviewed: 17-Oct-2022 / QC No. rroa-22-71450 / Revised: 24-Oct-2022 / Manuscript No. rroa-22-71450 / Published Date: 31-Oct-2022 DOI: 10.4172/2375-4338.1000328 QI No. / rroa-22-71450
Abstract
Determining the extent and degree of germ plasm diversity and genetic relationships among breeding materials is an important aid in crop improvement research strategies with an understanding that genetic variability is the base for crop improvement providing an opportunity for plant breeders to develop new and improved cultivars with desirable traits and it is a key to reliable and sustainable production of crops through breeding. It has been also confirmed that measuring the available genetic diversity of crops is important for effective evaluation and utilization of germ plasms to explore their variability so as to identify necessary agronomic traits. For eradicating the problem of rice production, the national rice breeding and genetics research program of the country is introducing and evaluating different rice germ plasms for their environmental adaptability and agronomic performance with increasing the crops’ genetic diversity. Likely, 100 upland rice genotypes were introduced and evaluated with 3 nationally released upland rice varieties as standard checks using an augmented-RCBD experimental design so as to assess and determine the extent and pattern of their genetic variability using cluster analysis bringing them into similar groups based on their important agronomic traits. Each genotype was planted on a plot area of 2.5 m2 involving 4 rows per plot with 0.25m spacing between each row. The seeds were drilled in rows with a seed rate of 60 kgha-1. Nitrogen-phosphorus-sulphur (NPS) and Urea fertilizers were applied in the amount of 124 kg ha-1 and 100 kgha-1 respectively. The quantitative traits such as days to 50% heading, days to 85% maturity, plant height, panicle length, number of filled grains per a panicle and number of unfilled grains per a panicle, grain yield and 1000 seed weight were measured from 97 genotypes since 6 of the genotypes were not emerged and data were not yet collected from them. The collected quantitative traits were subjected to clustering analysis using XLSTAT 5.03 statistical software so as to determine the extent and pattern of the genetic variability of 97 upland rice genotypes. During clustering analysis, the genotypes were grouped into five clusters with different Euclidian distances confirming the presence of genetic variability among the evaluated upland rice genotypes. The genotype with the highest grain (6298 kgha-1) yield was obtained and included under cluster-III.
Keywords: Cluster analysis; Upland rice genotype; Variability
Keywords
Cluster analysis; Upland rice genotype; Variability
Introduction
Rice (Oryza sativa L., 2n = 2x = 24) is the staple food for more than half of the world’s population and is increasingly important in Ethiopia for ensuring food security of the country though the crop is relatively new to the country, with its introduction in the 1970s [1,2]. Currently, it is estimated that the country is endowed with about 30 million ha of cultivated land, of which 5.6 million hectare is categorized as highly suitable for irrigated rice production but the current rice production area is only 85,288.87 hectares (CSA, 2021) [3]. However, the national rice breeding and genetics research program is introducing and evaluating different rice germ plasms targeting for their environmental adaptability and agronomic performance (high grain yield, tolerant for biotic and abiotic stresses and for other quality traits) aiming at enhancing the rice national productivity with increasing its annual production and area coverage.Deterring the extent and degree of germ plasm diversity and genetic relationships among breeding materials is a vital aid in crop improvement strategies with an understanding that genetic variability is the base for crop improvement providing the opportunity for plant breeders to develop new and improved cultivars with desirable characteristics and it is a key to reliable and sustainable production of the food crops through breeding [4]. It has been also confirmed that measuring the available diversity of crops is important for effective evaluation and utilization of germ plasms to explore their variability so as to identify desirable agronomic attributes [5]. Bringing improvements over the existing crop varieties is a continuous process in plant breeding there identifying diverse parents having high genetic variability for combining desirable characters and similarly the effectiveness of any rice improvement program depends on the utilization of different germ plasm from around the world [6]. Therefore, knowledge of sound genetic diversity is important for implementing any breeding program targeting crop improvement with exploring the variability present in their germ plasm for identification of desirable agronomic attributes [7, 8]. Similarly, to bring improvements over the rice varieties, introduction and evaluation of different rice germ plasms for different ecosystems to increase the national genetic diversity of the crop is one of the usual researches works of Ethiopian institute of agricultural research and a total of 3,336 germ plasms for rain fed upland, rain fed lowland and irrigated rice ecosystems have been introduced with the major objectives of high yielding, medium maturing, high biomass, white caryopsis, abiotic stress tolerance (cold and salinity), biotic resistance (blast, brown spot and sheath rot) rice varieties where the introduced germ plasms pass through a serious of evaluation stages for variety release and a total of almost 39 improved rice varieties has been released [9].
Still, the national demand for improved rice technologies is increasing from time to time that is why the national rice research program of the country collected and introduced a set of rice germ plasm from different external sources with the objective of characterizing and evaluating their environmental adaptability in lined with their genetic diversity and agronomic potential so as to identifying the promising rice genotypes for further rice improvement research program. With the same trend, 100 upland rice genotypes have been introduced to be involved in the rice germ plasm enhancement research sub component of the national rice research program of the country for being evaluated under the observation nursery trial at Pawe agricultural research center to assess their environmental adaptability and agronomic performances. In addition, this activity was also designed to determine the extent and pattern of the genetic and phenotypic variability of the upland rice genotypes with using cluster analysis bringing them into similarity groups based on important agronomic traits.
Materials and Methods
Description of the Experimental site
The trial was implemented at Pawe agricultural research center for one year during 2019/20 main cropping season under rain-fed condition (Figure 1).
The experimental area is located in the north western regions of Ethiopia at about 575km north west of Addis Ababa at a latitude of 11°19’N and longitude of 36°24’E and at an altitude of 1120mas which is characterized by hot to warm moist conditions with mean minimum and maximum temperature of 16oc and 32oc, respectively and has an average annual rainfall of 1587mm for five to seven months duration [10].
Plant Materials
Hundred upland rice genotypes with three standard checks (supplement-1) were used. The genotypes were collected and introduced from China and Japan in collaboration with IRRI and Africa-Rice research center.
Experimental Design and Procedures
An Augmented-RCBD experimental design was used with a plot area of 2.5m2 with 4 rows per plot, 0.5m and 1m spacing between plots and blocks respectively, an inter-row spacing of 0.25m and a seed rate of 60 kgha-1 was used. Inorganic fertilizers of NPS (Nitrogenphosphorus- sulphur containing fertilizer) and Urea with an amount of 124 kgha-1 and 100 kgha-1 respectively were applied. The total NPS was applied at planting, whereas the urea was used with three splits (1/3 at planting, 1/3 at tillering after weeding and 1/3 at the crop’s panicle initiation stage).
Data Collection and Statistical Analysis
A plot and plant based quantitative characteristics of rice were collected following the appropriate growth stage of the rice crop from each experimental unit. six newly tested genotypes failed to germinate and hence no data collection was done from the plots where those missed genotypes were planted. Thus, the quantitative data were collected from 97 upland rice genotypes. The traits such as days to 50% heading (DH), days to 85% maturity (DM), plant height in cm (PH), panicle length in cm (PL), number of filled grains per a central main panicle (NFG), number of unfilled grains per a central main panicle (NUFG), grain yield in kgh-1 and 1000 seed weight in gm (TSW) were considered and subjected to average linkage hierarchal clustering analysis using XLSTAT 5.03 statistical software [11]. Clustering analysis is very important to see-through complex relationships among populations of diverse origins in a more simplified manner [12]. Thus, the average linkage hierarchical clustering (AHC) analysis was computed to determine the extent and pattern of genetic and phenotypic diversity among the tested upland rice genotypes based on their corresponding traits. The closeness between two upland rice genotypes was measured by calculating at what point they are dissimilar using Euclidian distance (Ward’s method) while grouping 97 upland rice genotypes.
Results and Discussion
The descriptive statistical estimation of the traits
From the descriptive statistical analysis result of the measured quantitative traits of the upland rice genotypes, the average value of 131 days for 85% days to maturity (DM) was obtained indicating that there is a probability of getting a medium maturing upland rice genotypes which is compatible with the rain fed upland rice cultivation season of the testing location and other agro-ecologically similar areas of Ethiopia (Table 1).
| S/N | Agronomic traits | Min. Value | Max. Value | Mean Value | Std. dev |
|---|---|---|---|---|---|
| 1 | DH | 79 | 118 | 95 | 9.93 |
| 2 | DM | 117 | 158 | 131 | 6.75 |
| 3 | PH | 55.80 | 114.40 | 85.64 | 13.68 |
| 4 | PL | 16.20 | 23.00 | 20.96 | 1.94 |
| 5 | NFG | 67 | 172 | 105 | 20.73 |
| 6 | NUFG | 1 | 27 | 6 | 3.39 |
| 7 | TSW | 18.38 | 36.10 | 26.13 | 4.09 |
| 8 | GY | 1493.48 | 6298 | 3950.48 | 231.96 |
Where: DH-days to 50% heading; DM-days to 85% maturity PH-plant height in cm; PL-a panicle length of the main central tiller of a sampled upland rice plant in cm; NFG-number of filled grains per a central main panicle of the sampled upland rice plant; NUFG-number of unfilled grains per a central main panicle of the sampled upland rice plant; GY-grain yield in kg per hectare and TSW-1000 seed weight in gram
Table 1. The descriptive statistical analysis result of 8 quantitative traits collected from 97 upland rice genotypes tested at the research station of Pawe agricultural research center.
As it has been noted by IRRI (1984), standards of classifying the rice genotypes based on their time of physiological maturity, the genotypes could be classified as extra early maturing (up to 95 days), very early maturing (96-110 days), early maturing (111-125 days), medium maturing (126-140 days) and late maturing (141 days and above). Thus, 16 of the tested 97 upland rice genotypes could be classified as an early maturing genotype. 73 of the upland rice genotypes with days to maturity ranging from 126 to 140 days to 85% were identified as a medium maturing and 8 of the genotypes found to be late maturing with a number of 85% days to maturity ranging from 143 to 158 numbers of days to 85% maturity.
While considering their plant height (PH), the genotypes obtained values ranging from 55.80 to 114.40cm with an average panicle length (PL) of 20.96cm. For number of filled grain per a panicle (NFG), the average value of 105 grains (ranged from 67 to 172 grains) was obtained from which 29 upland rice genotypes scored more than the average value of NFG scored by the standard check called ‘Adet-1’ which scored a better value of 113 NFG ensuring the higher probability of getting the upland rice genotypes with a better expected grain yield. It was also found that almost 61% of the upland rice genotypes had in-determinant number of unfilled grains per a panicle which was less than or equal to the average value of NUFG (6 grains per a panicle). These two results for number of filled and unfilled grains per a panicle indicated that these genotypes had a higher predicted yield potential when compared to the best performing genotypes among the checks directing that they will have a greater chance to be subjected to further rice improvement program focusing on grain yield as the trait of interest.
The 1000 seed weight (TSW) of the tested upland rice genotypes were ranged from 18.38gm to 36.10gm with an average value of 26.13gm where 15 of the genotypes scored a better TSW when compared to a standard check called ‘Nerica-4’ with the higher TSW value of 30.27gm. A high TSW will increase germination percentage, seedling emergence, tillering efficiency, plant population density and grain yield [13]. Therefore, the higher TSW for 15 upland rice genotypes is an indication for the presence of most promising rice genotypes with a better genetic advantage over the best performing standard checks for their population stand, seedling vigor, tillering efficiency and yield potential.
Clustering the upland rice genotypes using eight quantitative traits
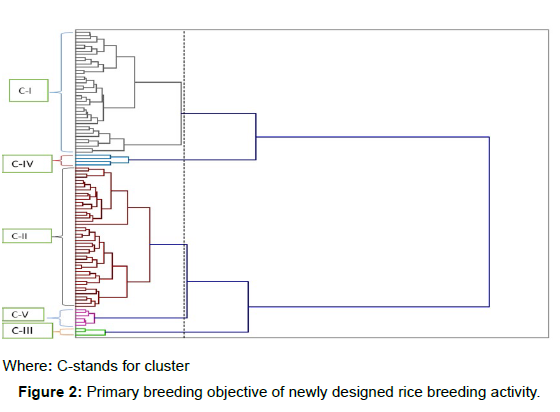
From the clustering result, the upland rice genotypes were grouped into five main clusters approving that there was a genetic variation among the tested rice genotypes with 86.39% of between cluster variance decomposition and 13.61% of a variance decomposition with in each cluster (cluster) (Table 2). Among the reported clusters, Cluster II contained the highest number of genotypes (45 genotypes) and cluster I contained the second large number of genotypes (36 genotypes) while, cluster III, cluster IV, cluster V contained 3,4 and 6 genotypes, respectively.
| Variance decomposition | Percent of Variance |
|---|---|
| Within-cluster | 13.61% |
| Between-clusters | 86.39% |
Table 2. Variance decomposition and extent of within and between-cluster variance
The cluster mean value of each measured trait in each cluster was computed for the tested upland rice genotypes. From the analysis result, an average cluster mean value of 85% days to maturity (DM) ranging from 129 to 134 days was obtained for genotypes grouped under cluster III and V respectively while clustering the 97 upland rice genotypes. This indicated that there is a greater chance of obtaining medium maturing genotypes which were grouped under all the five clusters.
The highest cluster mean values of plant height ((105cm)) was recorded among the genotype grouped under cluster III and the shortest cluster mean value of the trait (83.05cm) was obtained among the genotypes grouped under cluster IV.
Tall varieties (104 cm and taller) are suitable for flood-prone and unlevelled fields where lodging may be a problem. The rice varieties with a medium height (100 to 102 cm) are not susceptible to lodging and are suitable for most areas when fertilizer is used. Short varieties (less than 100 cm in height) are best suited to level fields especially in irrigated areas and are responsive to fertilizers [14]. Hence, most genotypes included under all clusters except cluster III had a plant height (PH) less than 100 cm indicating that there is indication for obtaining short upland rice genotypes which might be best suited to level fields especially in irrigated areas and will be more responsive to fertilizers. The highest cluster mean value of plant height (105 cm) for the upland rice genotypes grouped under III indicates the probability of obtaining a genotype which is suitable for flood-prone and unlevelled fields where lodging may be a problem. A highest cluster mean value of panicle length (21.30 cm) and number of filled grains per panicle (113 grains per panicle) with a higher cluster mean value for a grain yield (5286.72 kgha-1) was obtained for cluster V directing that those genotype with a better expected grain yield potential could be obtained from those tested upland rice genotypes clustered under this cluster.
In rice, yield could be determined by indirect traits like panicle length and grains per panicle as well as direct traits like filled grains per panicle and 1000-grain-weight [15]. Thus, the above results of the higher cluster mean values of panicle length (PL) and number of filled grains per panicle (NFG) for genotypes grouped under cluster V indicated that the respective rice genotypes could have a better expected grain yield potential when compared to those genotypes in other clusters though the highest cluster mean value of grain yield was recorded by those genotypes grouped under cluster III (6152.92 kgha-1). Therefore, a better attention could be given for those genotypes included under cluster-V and cluster-III while considering grain yield as the primary breeding objective of newly designed rice breeding activity (Figure 2) (Table 3) (Table 4).
| Cluster | Number of genotypes |
List of genotypes grouped under the assigned cluster number |
|---|---|---|
| I | 39 | G-1, G-2, G-8, G-14, G-17, G-18, G-19, G-20, G-22, G-24, G-25, G-26, G-28, G-31, G-32, G-38, G-40, G-41, G-42, G-44, G-55, G-56, G-57, G-58, G-59, G-60, G-61, G-65, G-67, G-69, G-78, G-79, G-81, G-86, G-89, G-91, G-93, G-94 and G-103 |
| II | 45 | G-3, G-4, G-5, G-9, G-11, G-12, G-13, G-15, G-16, G-27, G-30, G-33, G-35, G-36, G-37, G-39, G-43, G-45, G-47, G-48, G-49, G-50, G-51, G-52, G-54, G-62, G-63, G-64, G-66, G-68, G-70, G-71, G-72, G-75, G-76, G-77, G-80, G-82, G-84, G-85, G-87, G-88, G-90, G-101 and G-102 |
| III | 3 | G-6, G-7 and G-10 |
| IV | 4 | G-21, G-23, G-53 and G-92 |
| V | 6 | G-29, G-34, G-46, G-73, G-74 and G-83 |
Where: G-stands for genotype
Table 3. List of the upland rice genotypes grouped under each cluster
| Cluster/Traits | DH | DM | PH | PL | NFG | NUFG | TSW | GY |
|---|---|---|---|---|---|---|---|---|
| I | 94 | 132 | 84.55 | 20.97 | 105 | 6 | 25.43 | 3270.32 |
| II | 96 | 131 | 85.57 | 20.84 | 103 | 6 | 26.75 | 4413.40 |
| III | 95 | 134 | 105.00 | 22.80 | 115 | 5 | 26.02 | 6152.92 |
| IV | 99 | 131 | 83.05 | 20.23 | 104 | 9 | 25.16 | 1718.04 |
| V | 90 | 129 | 85.23 | 21.30 | 113 | 6 | 26.69 | 5286.72 |
Where: DH-days to 50% heading, DM-days to 85% maturity, PH-plant height in cm, PL-panicle length in cm, NFG-number of filled grains per a panicle, NUFG-number of unfilled grains per a panicle, GY-grain yield in kg per hectare and TSW-1000 seed weight in gram
Table 4. The mean values (Cluster centroids) of eight quantitative traits in each cluster of 97 upland rice genotypes
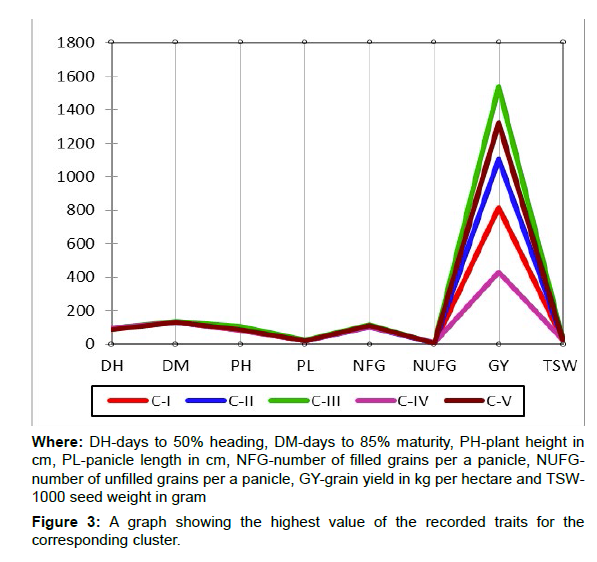
The upland rice genotype with the highest grain yield value was grouped under cluster III. (Figure 3)
Note: The unit of GY (grain yield) in this case is in gram per plot (the grain yield harvested from a plot size of 2.5m2. This has been done to make the graph well-structured and visible.
While computing the inter-cluster distances (distance between the cluster centroids for each measured quantitative trait dissimilarity) using Euclidean distance calculation, the maximum inter-cluster distance was obtained between cluster III and IV (1109.03) while clustering the tested upland rice genotypes. Inter cluster distance is the main criterion for selection of the genotypes implying that the genotypes belonging to the cluster with maximum inter cluster distance are genetically more divergent suggesting that selection of genotypes should be based upon large inter cluster distances, which may lead to broad spectrum of favorable genetic variability [16]. Therefore, it was confirmed that selection of genotypes for the improvement of traits of interest among the eight quantitative traits of the tested upland rice genotypes would be focused on those rice genotypes grouped under cluster III and IV (Table 5).
| Clusters | I | II | III | IV | V |
|---|---|---|---|---|---|
| I | 0 | 285.79 | 721.03 | 388.12 | 504.19 |
| II | 0 | 435.52 | 673.86 | 218.67 | |
| III | 0 | 1109.03 | 217.60 | ||
| IV | 0 | 892.28 | |||
| V | 0 | ||||
| Within-cluster variance | 9809.18 | 6668.28 | 1136.66 | 4284.55 | 482.26 |
| Intra-cluster distances | 86.85 | 74.68 | 25.54 | 54.93 | 19.36 |
Table 5. Inter-cluster distances (Euclidean distances) and intra-cluster distances for the clustering pattern of 97 upland rice genotypes and their variances with in a cluster
The higher average intra cluster distance was observed for Cluster I, II, and IV intra-cluster distances (86.85, 74.68 and 54.93 respectively) indicating that there was a high genetic diversity among the tested genotypes grouped with in these clusters. These clusters were also identified with a higher with in cluster variance of 9809.18, 6668.28 and 4284.55 for cluster I, II, and IV respectively confirming that there is a higher genetic diversity among the upland rice genotypes of the respective clusters. The highest inter cluster distance was observed between cluster. The lowest inter clusters distance was found between cluster III and V (217.60) and the highest distance was found between cluster III and IV (1109.03).
The genotypes grouped into the same cluster, as well as, the lowest values of inter cluster distance indicating a close relationship between them and exhibited the lowest divergence’s degree from one another giving a hint that no need of making hybridization between genotypes belonging to the same cluster or between genotypes from closely related clusters because no transgressive segregants are predicted from these combinations. In addition, the highest values of inter cluster distance suggests the maximum variability among the clusters or between different genotypes from two different clusters with the highest inter cluster distances from which hybridization programs could be designed in such a way that the parental genotypes relating to different clusters with high amount of divergence could be used to get appropriate transgressive segregants [17]. Thus, hybridization program could be designed by considering genotypes from cluster III and IV or from cluster I.
Conclusion
The clustering analysis where 97 upland rice genotypes group into five different clusters directs the presence of genetic diversity among the tested upland rice genotypes from which Cluster-I has the highest intra-cluster distance (86.85) confirming the presence of a highest genetic diversity among the genotypes included in this cluster. The highest value of inter cluster distance between cluster III and IV also suggests that there is the maximum variability between the upland rice genotypes grouped under these cluster. Thus, hybridization or other rice improvement programs could consider the genotypes from cluster III and IV or genotypes grouped under cluster I so as to get appropriate transgressive segregants of the traits of interest. The obtained cluster mean values of 85% days to maturity ranging from 129 for cluster-V to 134 for cluster-III indicates that there is a greater chance of obtaining medium maturing upland rice genotypes clustered under all the five clusters. The finding of upland rice genotypes with higher number of filled grains and lower number of unfilled grains per a central panicle, higher grain yield potential and with the higher 1000 seed weight values when compared to the best performing standard checks indicates that there is a greater chance of getting genotypes with a better genetic advantage over the best performing standard checks used during the experiment for their population stand, seedling vigor, tillering efficiency and yield potential. Thus, an attention could be given for those genotypes with a greater value of these quantitative traits while selecting genotypes to be improved for the desired traits of the genotypes to be implemented in the next breeding program and a value will be added on the rice research and development program of Ethiopia.
Acknowledgement
None
Conflict of Interest
None
References
- Bond N, Thomson J, Reich P and Stein J (2011).Using species distribution models to infer potential climate change-induced range shifts of freshwater fish in south-eastern Australia. Mar Freshw Res AU 62:1043-1061.
- Araújo M B, Pearson R G, Thuiller W, and Erhard M (2005). Validation of species–climate impact models under climate change. Glob Change Biol US 11:1504–1513.
- Gibson C, Meyer J, Poff N, Hay L, and Georgakakos A (2005). Flow regime alterations under changing climate in two river basins: implications for freshwater ecosystems. River Res Appl UK 21:849–864.
- Kearney M, Porter W (2009). Mechanistic niche modelling: combining physiological and spatial data to predict species’ ranges. Ecol Lett UK 12:334–350.
- Smakhtin V U (2001). Low flow hydrology: a review. J Hydrol EU 240:147–186.
- Kearney M, Porter W (2009). Mechanistic niche modelling: combining physiological and spatial data to predict species’ ranges. Ecol Lett UK 12:334–350.
- Smakhtin V U (2001). Low flow hydrology: a review. J Hydrol EU 240:147–186.
- Snyder R L, Melo-Abreu J P (2005) Frost protection: fundamentals, practice, and economics. FAO EU 1:1-72.
- Flannery K V (2008) Origins and ecological effects of early domestication in Iran and the Near East. IInd Edn Routledge UK:1-28.
- James M B(2001) The Hohokam of Southwest North America.J World Pre hist 15: 257–311.
- Siebert S J et al(2006) The Digital Global Map of Irrigation Areas – Development and Validation of Map Version 4.Germany EU.
- Frenken K (2005) Irrigation in Africa in figures – AQUASTAT Survey – 2005:Water Reports. FAO EU:1-649.
- Provenzano G (2007) Using HYDRUS-2D Simulation Model to Evaluate Wetted Soil Volume in Subsurface Drip Irrigation Systems. J Irrig Drain Eng US. 133: 342–350.
- Snyder R L, Melo-Abreu J P (2005) Frost protection: fundamentals, practice, and economics. FAO EU 1:1-72.
- Flannery K V (2008) Origins and ecological effects of early domestication in Iran and the Near East. IInd Edn Routledge UK:1-28.
- James M B(2001) The Hohokam of Southwest North America.J World Pre hist 15: 257–311.
- Siebert S J et al(2006) The Digital Global Map of Irrigation Areas – Development and Validation of Map Version 4.Germany EU.
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, GoogleScholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Citation: Muchie GG, Abebe D (2022) Determining the Level of Genotypic Variability of Different Upland Rice Genotypes Using Cluster Analysis. J Rice Res 10: 328. DOI: 10.4172/2375-4338.1000328
Copyright: © 2022 Muchie GG. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 2025
- [From(publication date): 0-2022 - Sep 23, 2025]
- Breakdown by view type
- HTML page views: 1613
- PDF downloads: 412