Detection of SARS-CoV-2 in Saliva Using Tailed Amplicon Sequencing
Received: 08-Jun-2021 / Accepted Date: 22-Jun-2021 / Published Date: 29-Jun-2021 DOI: 10.4172/2332-0877.1000464
Abstract
The most recent virus from the Coronaviridae family infecting humans, SARS-CoV-2, has resulted in a global pandemic. As part of the surveillance efforts, SARS-CoV-2 genomes are increasingly being made publicly available. Methods that include both short- and long-read sequencing have been used to elucidate SARS-CoV-2 genomes; however, many of these untargeted approaches may require deeper sequencing for greater genome coverage. For this reason, sequence capture or amplicon-based approaches for SARS-CoV-2 genome sequencing have been developed. The present proof-of-concept study evaluated a modified sequence capture approach, namely, tailed amplicon sequencing, to determine SARS-CoV-2 near complete genome sequences from the saliva of infected individuals. Particularly, the suitability of saliva samples stored at room temperature using OMNIgene® •ORAL OME-505 was evaluated. The tailed amplicon sequencing approach poses the additional advantage of being a cost-effective method for library preparation. Different known SARS-CoV-2 variants were identified across the infected subjects, with an average of >99.4% genome coverage. This methodology also enabled robust genomic surveillance using phylogenetic analyses. The present study supports the suitability of saliva stored at room temperature using collection devices for SARS-CoV-2 variant detection. Importantly, the present study supports the use of tailed amplicon sequencing approaches as an alternative, cost-effective method for SARS-CoV-2 detection in saliva for genomic surveillance.
Keywords: Genomic surveillance; SARS-CoV-2; Sequence capture; Tailed amplicon sequencing
Introduction
Human saliva is home to over 700 microbial species, and serves as protection against bacterial, fungal and viral infections that could potentially reach the respiratory tract [1]. It is composed of water and the secretion of salivary glands, dental plaque, as well as nasal and bronchial secretions; thus, several different viruses in human saliva can be identified, some of which may cause disease. The interactions with viruses and specific saliva components can be virus-specific, complex and can potentially influence their detection as a result of the biological functions of saliva [2].
Viruses including, but not limited to hepatitis A [3] hepatitis B [4,5] cytomegalovirus [6], Epstein Barr virus [7], Zika virus [8], and Severe Acute Respiratory Syndrome-Associated coronavirus (SARS-CoV) [9] have all been detected in the saliva of infected individuals. This has prompted the investigation of saliva as a potential sample type for the diagnosis of several of the mentioned viruses, and of potential emerging and re-emerging viral pathogens.
The most recent virus from the Coronaviridae family known to infect humans, SARS-CoV-2 has also been identified in the saliva of infected individuals [2]. While the origin of SARS-CoV-2 is a subject of ongoing research and speculation, it shares 96% identity with a bat coronavirus [10], 90% with coronaviruses present in pangolins [11] and 80% with SARS-CoV. SARS-CoV-2 is known to also be transmitted through aerosol droplets, which may also include salivary droplets. Several studies have identified SARS-CoV-2 in saliva [12,2], and have proposed its use as an alternative to Nasopharyngeal (NP) and Oropharyngeal (OP) sample collection, which pose additional discomfort and the need for trained personnel [13].
In addition, with the availability of at-home collection kits, subjects are more willing to self-collect saliva [14]. Supervised, self-collected saliva has shown to perform similarly to clinician-collected NP swabs for the detection of SARS-CoV-2 in terms of virus detection and quantification [15]. Success of detection methods for SARS-CoV-2 depends, among many things, on the persistence and inactivation of the virus and nucleic acids.
For example, SARS-CoV-2 is detectable using Reverse Transcription PCR (RT-PCR) for an average of 18 to 20 days, and up 21 to 26 days in some instances [16,13], Saliva has also been suitable for antibody testing, showing that SARS-CoV-2 antibodies can be detected as early as 10 days. This further supports the use of saliva for the detection of SARS-CoV-2 nucleic acids and antibodies. Other factors affecting SARS-CoV-2 detection include efficient viral lysis, with various commercially available kits and methods showing varying levels of efficiency [17].
The gold standard for the detection of SARS-CoV-2 in any sample type is RT-PCR or quantitative RT-PCR (RT-qPCR) [18]. As with any PCR-based method, success will depend on primer specificity; thus, the RNA target genome(s) sequence(s) should be known in order to increase sensitivity [19]. Other, less evaluated methods for the detection and surveillance of SARS-CoV-2 in various sample types include high-throughput sequencing. RNA high-throughput sequencing was originally used in combination with other methods to unknown origin, showing a similarity close to 90% to coronaviruses identify the novel coronavirus in subjects suffering from pneumonia of present in bats [20]. High throughput sequencing can continue to be used for the surveillance of known and novel SARS-CoV-2 variants, and also to determine genetic diversity, which is usually not provided by RT-PCR or RT-qPCR [21].
Two approaches have been developed for the detection of SARS-CoV- 2 in various sample types using high-throughput sequencing, namely, untargeted and amplicon-based. Untargeted high-throughput sequencing provides the advantage of detecting and monitoring both known and emerging SARS-CoV-2 variants, with the caveat of requiring deeper sequencing to obtain the needed genome coverage for identification, making this approach relatively more cost-prohibiting. Amplicon high-throughput sequencing, or sequence capture methods, has been more widely applied for the detection of SARS-CoV-2 as it represents a more cost-effective approach. One caveat of sequence capture approaches is the failure to cover the entire viral genome as the primers usually cannot cover the genomic ends [22].
Approaches, such as the ARTIC network, have developed methods for amplicon pool preparation for the sequencing of SARS-CoV-2 which involve a ‘lab-in-a-suitcase’ that can be used in remote and resource-limited areas. In the ARTIC network protocol, the first cDNA strand is enriched by amplifying with two different pools of primers. This generates amplicons tiling the virus genome, which are then subjected to either Illumina or Oxford Nanopore library preparation and sequencing. More recently, a modification to the ARTIC network protocol, known as tailed amplicon sequencing, has been developed to reduce library preparation cost and time [22].
The method has been shown to achieve results comparable to the ARTIC network protocol [22]. Briefly, in the tailed amplicon approach, the first cDNA strand is enriched using the ARTIC v3 primers. In this case, the primers also contain adapter tails that allow sequencing libraries to be created through a second indexing PCR. This, in turn, adds sample-specific barcodes and flow cell adapters. Notably, this modification to the ARTIC protocol has not been extensively tested in saliva samples. Therefore, the main aim was to evaluate the suitability of the modified tailed amplicon sequencing protocol for the detection of SARS-CoV-2 and its ability to discern potential variants in human saliva.
Materials and Methods
Sample collection and RNA extraction
Saliva samples were collected from various geographical locations in United States using the OMNIgene® •ORAL OME-505 device (DNA Genotek, Inc) and shipped to Clinical Reference Laboratory, Inc. for testing using the CRL Rapid ResponseTM COVID-19 Test. After testing, the remaining saliva samples collected and stabilized in the OMNI gene® •ORAL OME-505 devices, were stored at room temperature for up to 15 days prior to RNA extraction (Table 1).
| Saliva sample ID | Pangolin Lineage | Nextclade Clade | Collection Location | Diagnosis Ct value | Days from sample collection to RNA extraction | Fast qc Raw Read Pairs | Depth after trimming | Coverage (%) After Trimming | ivar num variants identified |
|---|---|---|---|---|---|---|---|---|---|
| A | B.1.2 | 20G | Belton, MO | 27.01 | 15 | 192151 | 2316.21 | 99.96 | 18 |
| B | B.1 | 20C | Wilmington, DE | 14.01 | 12 | 260188 | 2413.47 | 99.44 | 21 |
| C | B.1.2 | 20G | Bronx, NY | 16.5 | 3 | 233789 | 2547.39 | 99.73 | 29 |
| D | B.1.1.222 | 20B | Blacklick, OH | 17.4 | 7 | 137894 | 1571.84 | 99.96 | 29 |
| E | P.2 | 20B | Orient, OH | 17.64 | 11 | 166102 | 2166.38 | 99.76 | 27 |
| F | B.1.427 | 20C | Manhattan, KS | 17.9 | 4 | 135078 | 1527.5 | 99.91 | 30 |
| G | B.1.2 | 20G | Carson, CA | 18.28 | 2 | 212666 | 2146.3 | 99.81 | 22 |
| H | B.1.1.7 | 20I/501Y.V1 | Topeka, KS | 18.55 | 7 | 202621 | 2527.93 | 99.83 | 39 |
Table 1: Summary of relevant metadata associated with saliva samples including: Pangolin lineage which indicates the strain identity, Nexclade clade identification which shows the phylogenetic lineage of the variant, qPCR Ct value for diagnosis, number of raw fastq reads, read depth on the genome after trimming primers, percent coverage of the genome after trimming and the number of variants (mutations) identified using iVar.
At a later point, one to fifteen days post diagnosis; eight randomly selected samples from a pool of samples which had previously tested positive for SARS-CoV-2 by the CRL Rapid ResponseTM COVID-19 Test were selected to be sequenced. The only criteria used for sample selection was an initial Ct value <30 (Table 1). The randomly selected samples were extracted using the Zymo Quick RNA/DNA Viral Mag Bead kit (Cat. No. R2141) following manufacturer’s instructions.
Sample processing, sequencing and bioinformatic analyses
The extracted RNA was then prepared and sequenced using previously described methods in [22]. Briefly, the integrity of the extracted RNA was analyzed as described previously. After RNA quality and integrity was checked, RT-qPCR was also performed as described previously. RNA was also processed through the amplicon-based sequencing method that utilizes adapter tails with the ARTIC network v3 primers, allowing for a more efficient library preparation [7].
Sequencing was also performed as previously described using a Mi Seq 600 cycle v3 kit following manufacturer’s instructions. After sequencing, the paired ends were joined using PANDA seq [23]. Unaligned reads were aligned to Wuhan-Hu-1 SARS-CoV-2 genome (MN908947.3) using BWA [24,20]. The Ivar software package was used for trimming and filtering reads [25]. Ivar was also used to call variants and generate consensus sequences.
The consensus sequences were then strain typed using Pangolin (github.com/cov-lineages/pangolin). Genome coverage plots were created using custom R scripts. Phylogenetic tree was created using the Next strain web interface [26,27] (v0.14.2, commit: f62d906, build 655).
Results and Discussion
The present proof-of-concept study evaluated a modified sequence capture approach, namely tailed amplicon sequencing, for the detection of SARS-CoV-2 in the saliva of infected individuals [21]. Detection of SARS-CoV-2 in the saliva of infected individuals, both symptomatic and asymptomatic, has expanded the toolbox of methods for the detection and diagnostics of the virus. While RT-PCR and RTqPCR are the standard methods for the detection of SARS-CoV-2 in various sample types, tailed amplicon sequencing approaches are also capable of identifying SARS-CoV-2 in the saliva of infected individuals. Notably, results from the present study showed that the tailed amplicon sequencing approach was successful in the identification of various SARS-CoV-2 variants in human saliva (Table 1). Results also showed the suitability of OMNI gene® •ORAL OME-505 for sufficient SARS-CoV-2 RNA recovery for tailed amplicon sequencing. Moreover, storage of saliva samples at room temperature using OMNI gene® •ORAL OME-505 for up to 15 days further supports the capacity of this collection device to capture saliva composition at the time of collection, essential for SARS-CoV-2 diagnostics and surveillance. While upper and lower respiratory tract specimens were first to be recommended for SARS-CoV-2 diagnosis, saliva has gained acceptance as a suitable, non-invasive sample type. Indeed, saliva has been included in numerous FDA Emergency Use Authorizations for the purpose of SARS-CoV-2 diagnosis, including CRL Rapid ResponseTM. The OMNI gene® •ORAL OME-505 saliva collection device, which has received EUA for SARS-CoV-2 sample collection, allows for self-collection and to circumvent the need to store samples on ice or frozen.
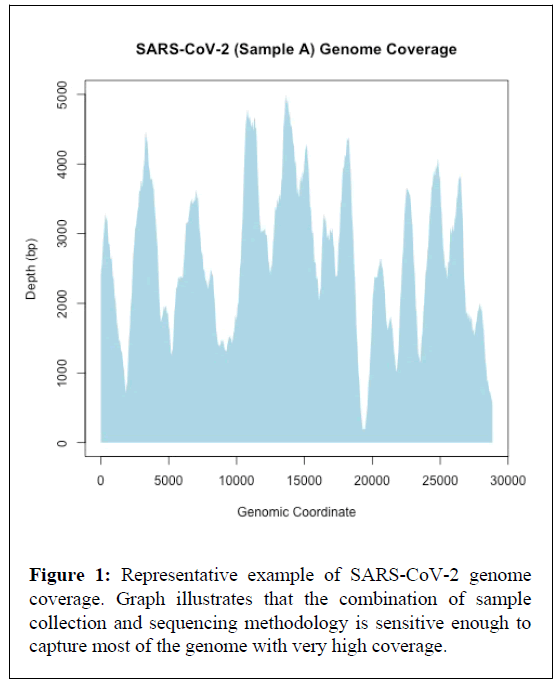
Interestingly, the tailed amplicon sequencing approach tested in the present study provided near complete genomes (>99.4% average genome coverage) for all eight samples tested (Table 1 and Figure 1). In addition, the tailed amplicon method produced a relatively uneven genome coverage balance. Indeed, this has been noted previously when comparing the tailed amplicon method with the ARTIC network protocol [22]. One feasible explanation for this unevenness in genome coverage is the better balance of the untailed primers utilized in the ARTIC network protocol.
Nevertheless, near complete SARS-CoV-2 genomes were obtained in the present study, which may improve insights into virus mutations, evolution, and adaptation (e.g., increased transmissibility and infectivity) compared to evolutionary relationships across spike protein sequences alone. Unlike specific genes, complete or near complete genomic sequences provide the most high-resolution information that allows determination of variant and strain relatedness during outbreaks and pandemics.
Thus, the development of cost-effective and less time-consuming protocols to determine genomic sequences is of importance. In addition, near complete genome information may also aid in the classification of SARS-CoV-2 variants into strains. SARS-CoV-2 strain level resolution using tailed amplicon sequencing approaches could potentially aid to bridge mutations within SARS-CoV-2 genomes at a global scale, which in turn may aid to understand virus transmission and population dynamics.
Variant level resolution of SARS-CoV-2 may also facilitate the identification of novel target regions for vaccine development and therapeutics, particularly regions across SARS-CoV-2 genomes that may be shared across variants and potential strains.
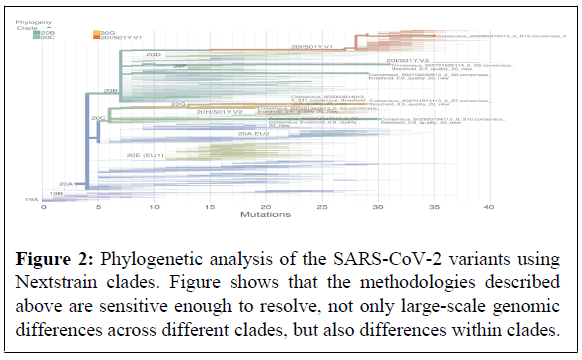
Near complete genomic sequences obtained using the tailed amplicon sequencing enabled deciphering phylogenetic relationships across the various SARS-CoV-2 variants identified in the saliva of the infected individuals in comparison with known clades (Figure 2). Results show that, overall, the tailed amplicon method described above is sensitive enough to resolve, not only large-scale genomic differences across different clades, but also differences within clades. Phylogenetic relatedness may further contribute to the understanding of the pathogenic dynamics of SARS-CoV-2 and associated strains over time and link these genetic variations to specific geographical regions, which in turn can target genomic surveillance efforts.
Conclusion
The present study evaluated tailed amplicon sequencing as a suitable approach for the detection of SARS-CoV-2 variants in the saliva of infected individuals collected using OMNIgene® •ORAL OME-505. Near complete genome sequences were obtained using the evaluated method, which in turn facilitated phylogenetic analysis with various SARS-CoV-2 variants. Near complete or complete genome sequences from SARS-CoV-2 variants continue to be invaluable in disease control and prevention efforts during the COVID-19 pandemic; thus, the evaluation of time- and cost-effective methods such as tailed amplicon sequencing is essential.
Author Contributions
AG-data analysis, figure generation and manuscript draft editing. TSR-writing and editing of original manuscript draft. HLF-sample collection and screening for positive samples RI-conceptualization and manuscript draft editing.
Acknowledgements
We thank Mike Tayeb, Geoffrey M. Graham and Austin Udocor for reviewing the manuscript draft.
Funding
No funding was received for this study.
Informed Consent and Release Statement
Written informed consent and release statement has been obtained from the patient(s) to publish this paper.
Institutional Review Board Statement
The study is exempt under 45 CFR § 46.104(d)(4), because the research involves the use of identifiable private information/bio specimens; and information, which may include information about bio specimens, is recorded by the investigator in such a manner that the identity of the human subjects cannot readily by ascertained directly or through identifiers linked to the subjects, the investigator does not contact the subjects, and the investigator will not re-identify subjects.
Conflict of Interest
AG and TSR are current employees of Diversigen Inc. RI is a current employee of DNA Genotek.
Data Availability
SARS-CoV-2 sequence genomes are available on the Global Initiative on Sharing Avian Influenza Data (GISAID) site under IDs EPI_ISL_2156824, EPI_ISL_2156826, EPI_ISL_2156827, EPI_ISL_2156821, EPI_ISL_2156810, EPI_ISL_2156811, EPI_ISL_2156823 and EPI_ISL_2156808.
References
- Kilian M, Chapple ILC, Hannig M, Marsh PD, Meuric V,et al. (2016) The oral microbiome - An update for oral healthcare professionals. Br Dent J 221:657-666.
- Li Y, Ren B, Peng X, Hu T, Li J,et al. (2020) Saliva is a non-negligible factor in the spread of COVID-19. Mol Oral Microbiol 35:141-145.
- Leon LAA, De Almeida AJ, De Paula VS, Tourinho RS, Villela DAM,et al. (2015)Longitudinal Study of Hepatitis A Infection by Saliva Sampling: The Kinetics of HAV Markers in Saliva Revealed the Application of Saliva Tests for Hepatitis A Study. PLoS One 10:e0145454.
- Khadse S V, Bajaj G, Vibhakar P, Nainani P, Ahuja R,et al. (2016) Evaluation of specificity and sensitivity of oral fluid for diagnosis of hepatitis B. J Clin Diagnostic Res 10:12-14.
- Parizad Elaheh Gholami, Parizad Eskandar Gholami, Khosravi A, Amraei M, Valizadeh A,et at. (2016) Comparing HBV viral load in serum, Cerumen, and saliva and correlation with HBeAg serum status in patients with chronic hepatitis B infection. Hepat Mon 16:30385.
- De Carvalho Cardoso ES, De Santos Jesus BL, da Silva Gomes LG, Sousa SMB, Gadelha SR,et al. (2015) The use of saliva as a practical and feasible alternative to urine in large-scale screening for congenital cytomegalovirus Han P, Ivanovski S. Saliva-friend and foe in the COVID-19 outbreak. Diagnostics 48:206-207.
- Kwok H, Chan KW, Chan KH, Chiang AKS (2015) Distribution, persistence and interchange of epstein-barr virus strains among PBMC, plasma and saliva of primary infection subjects. PLoS One 10:e0120710.
- Bonaldo MC, Ribeiro IP, Lima NS, Dos Santos AA, Menezes LS, et al. (2016) Isolation of infective Zika virus from urine and saliva of patients in Brazil. PLOS Negl Trop Dis 10:4816.
- Wang WK, Chen SY, Liu IJ, Chen YC, Chen HL,et al. (2004) Detection of SARS-associated coronavirus in throat wash and saliva in early diagnosis. Emerg Infect Dis 10:1213-1219.
- Zhou P, Yang X Lou, Wang XG, Hu B, Zhang L,et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature.
- Zhang T, Wu Q, Zhang Z (2020) Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr Biol 30:1578.
- Han P, Ivanovski S (2020) Saliva-friend and foe in the COVID-19 outbreak. Diagnostics 10:290.
- Yoon JG, Yoon J, Song JY, Yoon SY, Lim CS,et al.(2020) Clinical significance of a high SARS-CoV-2 viral load in the Saliva. J Korean Med Sci 35:e195.
- Valentine-Graves M, Hall E, Guest JL, Adam E, Valencia R,et al. (2020) At-home self-collection of saliva, oropharyngeal swabs and dried blood spots for sars-cov-2 diagnosis and serology: Postcollection acceptability of specimen collection process and patient confidence in specimens. PLoS One 15:e0236775.
- Noah K, Fred T, Vlad S, Agatha B, Laura D,et al. (2020) Self-collected oral fluid and nasal swabs demonstrate comparable sensitivity to clinician collected nasopharyngeal swabs for Covid-19 detection. MedRxiv 19:1589.
- To KKW, Tsang OTY, Leung WS, Tam AR, Wu TC,et al. (2020) Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect Dis 20:565-574.
- Chu AWH, Chan WM, Ip JD, Yip CCY, Chan JFW, et al. (2020) Evaluation of simple nucleic acid extraction methods for the detection of SARS-CoV-2 in nasopharyngeal and saliva specimens during global shortage of extraction kits. J Clin Virol 129:104519.
- Takeuchi Y, Furuchi M, Kamimoto A, Honda K, Matsumura H,et at. (2020) Saliva-based pcr tests for sars-cov-2 detection. J Oral Sci 62:350-351.
- Li D, Zhang J, Li J (2020) Primer design for quantitative real-time PCR for the emerging Coronavirus SARS-CoV-2. Theranostics 10:7150-7162.
- Wu F, Zhao S, Yu B, Chen YM, Wang W,et al. (2020) A new coronavirus associated with human respiratory disease in China. Nature 579:350-351.
- Pérez Cataluña A, Chiner-Oms Ã, Cuevas Ferrando E, DÃaz-Reolid A, Falcó I, et al. (2021) Detection Of Genomic Variants Of SARS-CoV-2 Circulating In Wastewater By High-Throughput Sequencing.
- Gohl DM, Garbe J, Grady P, Daniel J, Watson RHB, et al.(2020) A rapid, cost-effective tailed amplicon method for sequencing SARS-CoV-2. BMC Genomics 21.
- Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD,et at. (2012) Paired-end assembler for illumina sequences. BMC Bioinformatics 13:1-31.
- Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 20:589-595.
- Grubaugh ND, Gangavarapu K, Quick J, Matteson NL, De Jesus JG,et al. (2019) An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol 20.
- Hadfield J, Megill C, Bell SM, Huddleston J, Potter B,et al. (2018) NextStrain: Real-time tracking of pathogen evolution. Bioinformatics 34:4121-4123.
- Itokawa K, Sekizuka T, Hashino M, Tanaka R, Kuroda M (2020) A proposal of alternative primers for the ARTIC Network’s multiplex PCR to improve coverage of SARS-CoV-2 genome sequencing. BioRxiv 15:0239403
Citation: Garoutte A, Santiago-Rodriguez TM, Fehling HL, Iwasiow R (2021) Detection of SARS-CoV-2 in Saliva Using Tailed Amplicon Sequencing. J Infect Dis Ther 9:464. DOI: 10.4172/2332-0877.1000464
Copyright: © 2021 Garoutte A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 1930
- [From(publication date): 0-2021 - Nov 21, 2024]
- Breakdown by view type
- HTML page views: 1319
- PDF downloads: 611