Research Article Open Access
Detection of High Prevalence of TEM/SHV/CTX-M Genes in ESBL Producing and Multidrug Resistant Klebsiella Pneumoniae and Klebsiella Oxytoca
Diagbouga S1,2*, Salah F D1, Sadji A Y3, Dabire A M1, Nadembega C1, Kere A B3, Soubeiga S T1, Ouattara A K1, Zohoncon T1, Belemgnegre M1, Karou S4, and Simpore J11Department of Biochemistry-Microbiology, University of Ouaga I Prof Joseph Ki-Zerbo, BP 364 Ouagadougou, Burkina Faso
2Research Institute for Health Sciences (IRSS), 7192, Ouagadougou, Burkina Faso
3Bacteriology Laboratory, National Institute of Hygiene (INH), 1396, Lomé, Togo
4School of Biological and Food Technique (ESTBA), University of Lomé, 1515, Togo
- Corresponding Author:
- Serge Diagbouga
Department of Biochemistry-Microbiology
University of Ouaga I Prof Joseph Ki-Zerbo
BP 364 Ouagadougou, Burkina Faso
Tel: 00226 70 23 17 96
E-mail: diagbouga.serge@gmail.com
Received Date: August 19, 2016; Accepted Date: October 04, 2016; Published Date: October 08, 2016
Citation: Diagbouga S, Salah FD, Sadji AY, Dabire AM, Nadembega C, et al. (2016) Detection of High Prevalence of TEM/SHV/CTX-M Genes in ESBL Producing and Multidrug Resistant Klebsiella pneumoniae and Klebsiella Oxytoca. J Clin Diagn Res 4:130. doi: 10.4172/2376-0311.1000130
Copyright: © 2016, Diagbouga S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at JBR Journal of Clinical Diagnosis and Research
Abstract
Background: Klebsiella spp. are Enterobacteriaceae frequently isolated from pathological specimens during urinary tract infections, bloodstream infection, and pus. They are becoming more and more resistant to antibiotics and challenging treatment options. β-lactamases are a great variety of enzymes capable of inducing resistance to β- lactams. The objective of this study was to identify extended-spectrum-β-lactamase (ESBL) genes in Klebsiella spp. strains isolated from various specimens in Lomé, Togo. Methods: Sixty-four strains of Klebsiella spp. were isolated from different pathological specimens. They were then further characterized and tested against 3rd generation cephalosporin (ceftazidime, ceftriaxone, cefotaxime) and aztreonam. The detection of blaTEM, blaSHV and blaCTX-M was performed on these strains using simplex and multiplex PCR techniques. Results: Fifty five (85.94%) Klebsiella pneumoniae and 9 (14.06%) Klebsiella oxytoca were isolated. These strains derived from urine (n=33; 51.56%), vaginal swabs (n=21; 32.81%), pus (n=8; 12.5%) and sperm samples (n=2; 3.13%). All strains were resistant to cefepime. The resistance rate to other β-lactams was 29.69% (19/64) for piperacillin-tazobactam, 23.08% (12/52) for cefoxitin and 1.56% (1/64) for imipenem. Other inactive antibiotics were trimethoprim-sulfamethoxazole 96.72% (59/61), doxycycline 92.06% (58/63), ciprofloxacin 90.63% (58/64), nalidixic acid 80.95% (51/63), chloramphenicol 77.42% (48/62) and gentamicin 76.69% (51/64). Amikacin and fosfomycin remained the most active antibiotics with 1.56% (1/64) and 4.69% (3/64) resistance rates respectively. ESBL genes were detected in 63/64 (98.44%) strains. TEM/SHV/CTX-M was predominant 61.90% (39/63) followed by TEM/SHV 20.63% (13/63), SHV/CTX-M 11.11% (7/63), TEM/SHV 4.76% (3/63) and TEM 1.59% (1/63). Conclusion: ESBL genes occur more by combination 96.88% (62/64) than singularly 1.59% (1/63). These strains were also very resistant to quinolones and trimethoprim-sulfamethoxazole. These findings are of high importance in a medical and scientific perspective and may motivate decision makers towards a better monitoring and control of antimicrobial resistance in Togo.
Keywords
Klebsiella spp; Antimicrobial resistance; ESBL; TEM; SHV; CTX-M; Togo
Introduction
β-lactamases are serine enzymes (class A, C or D of Ambler classification) or metalloenzyme (Class B of Ambler classification) that confer resistance to β-lactams by hydrolyzing their β-lactam ring [1]. The discovery and use of new classes of β-lactams have immediately been followed by the emergence of new β-lactamases. The first reported β-lactamases TEM-1/2 and SHV-1, described since 1960 from E. coli and Klebsiella pneumoniae were usually chromosomal. They are able to inactivate penicillins (amoxicillin, ampicillin, ticarcillin) hence the name penicillinases narrow spectrum [2]. From the 80s, plasmid mediated extended-spectrum-β-lactamases (ESBLs) have been described and confer resistance to penicillins, oxyimino cephalosporins (cefotaxime, ceftazidime, ceftriaxone, cefuroxime, cefepime) and monobactams (aztreonam) [3-5]. These ESBLs are divided into several groups; the main ones are the derivated of TEM and SHV groups and CTX-M [6]. In recent years, the CTX-M ESBL spread around the world and have been described in the commensal flora and also in hospital-acquired and community infections with a tendency to supplant the first ESBL groups TEM and SHV [7-14].
Antibiotic resistance thus is a public health concern because of the difficulty to find treatment options and the fatal outcome to which they may lead [15-19]. Klebsiella spp. is one of the most frequently isolated Gram-negative bacteria in hospital and community infections with ESBL [20-22]. These ubiquitous bacteria, commensal of nasopharynx and gastrointestinal tract are responsible of community-acquired pneumonia, urinary tract infections, nosocomial infections, rhinoscleroma, ozena, chronic ulceration of the genital tract and colonization [23,24]. Klebsiella pneumoniae and Klebsiella oxytoca are the most medically important species, often isolated from severe infections in hospitalized or ambulatory patients [25-29]. Klebsiella spp. are naturally resistant to penicillins (amoxicillin, ampicillin, piperacillin and ticarcillin) due to a low production of a class A chromosomal β-lactamase (penicillinase). The severity of Klebsiella spp. infections is increased when the clinical strains acquire resistance genes, such as ESBLs genes, following the misuse of antibiotics and other risk factors such as a long hospitalization associated with regular antibiotics, poor adherence, and low socioeconomic conditions [30-32]. Previous studies have shown high frequency of ESBL in hospitals among Klebsiella spp. with variation from one continent to another. The global multi-center surveillance study Tigecycline Evaluation and Surveillance Trial (TEST) reported from 2004 to 2009 an overall prevalence of ESBL producing Klebsiella pneumoniae isolated from intensive care units (ICUs), ranging from 12.8% in North America to 26.6% in Europe, 33.8% in the Middle East, 35.6% in the Asia-Pacific region, 45.5% in Latin America an up to 54.9% in Africa [33]. In addition, Hoban et al. reported a prevalence of ESBL of 8.8% and 38.9% in North America and Europe respectively with a predominance of CTX-M15 subgroup [34] on Klebsiella pneumoniae strains isolated from urinary tract infections.
In Togo, the prevalence of ESBL was previously reported [35]; however, their molecular characterization was not yet described. However, a recent study focused on the molecular characterization of ESBL producing E. coli (Salah et al. submitted for publication). The present study aimed to characterize ESBL genes in Klebsiella spp. isolated from various pathological specimens received at the National Institute of Hygiene in Lomé, Togo.
Materials and Methods
Collection and identification of Klebsiella spp. strains, antimicrobial susceptibility testing, and detection of ESBL phenotype
Klebsiella spp. strains were collected during a prospective study from May 2013 to July 2015 in the bacteriology laboratory of the National Institute of Hygiene (INH) in Lomé, Togo. This institute is a public health service specialized in biomedical analysis, epidemiological surveillance, immunization, water, and food quality control. The strains were isolated from various pathological specimens including urine, vaginal swabs, pus, and sperm samples. The standard microbiological methods were used to isolate and purify bacterial strains on Mac-Conkey or eosin methylene blue (EMB) media. The strains were identified by the API 20E identification system (Biomérieux, Marcy-Etoile, France).
Antimicrobial susceptibility testing was performed and interpreted according to the 2014 recommendations of Antibiogram Committee of the French Society of Microbiology [36]. Antibiotics were purchased from BioRad (Marnes-la-Coquette, France) and included amoxicillin +clavulanate (AMC, 20/10 μg), piperacillin-tazobactam (TZP, 75/10 μg), cefoxitin (FOX, 30 μg), ceftriaxone (CRO, 30 μg), ceftazidime (CAZ, 30 μg), cefotaxime (CTX, 30 μg), cefepime (FEP, 30 μg), aztreonam (ATM, 30 μg), imipenem (IPM, 10 μg), amikacin (AMK, 30 μg), gentamicin (GEN, 15 μg), nalidixic acid (NAL, 30 μg) ciprofloxacin (CIP, 5 μg), trimethoprim-sulfamethoxazole (SXT 1.25/23,75 μg), fosfomycin (FOF, 50 μg), doxycycline (DOX, 30 μg) and chloramphenicol (CHL, 30 μg). The double disk synergy test was used to detect ESBL production [37]. E. coli ATCC 25922 was used as a control strain for antimicrobial susceptibility testing.
Klebsiella pneumoniae and Klebsiella oxytoca strains resistant to at least one 3rd generation cephalosporin, ceftazidime, ceftriaxone, cefotaxime, or to aztreonam were collected, stored in a storage medium (tryptic soy broth TSB) at -80°C. They were then transferred to the Molecular Biology Laboratory of CERBA/LABIOGENE in Ouagadougou, Burkina Faso for ESBL genes detection.
Extraction of bacterial DNA
The bacterial chromosomal and plasmid DNA was extracted by a boiling method. Briefly, from the TSB, strains were reactivated on tryptic soy agar (TSA) for 18-24 h and then inoculated in Luria Bertani broth (LB, 2 ml). After 18-24 h culture, LB broth were centrifuged (10 000 RPM/min for 10 min) and bacterial cells were suspended in 500 μl of phosphate buffer (100 mM, pH 7) to weaken the membranes. An immersion in a boiling water bath (100°C for 15 min) releases the genetic material. The DNA is then precipitated with 250 μl of absolute alcohol, washed twice in 1000 μl of 70% alcohol (stored at -20°C), dried and re-suspended in 100 μl of sterile water.
Detection of ESBL genes
The ESBL genes, blaTEM (TEM-1/2), blaSHV (SHV-1), blaCTX-MG1 (CTX-M1, 3 and 15), blaCTX-M-G2 and blaCTX-M-G9 (CTX-M9 and CTX-M14) were detected by PCR using the thermal cycler GeneAmp PCR System 9700 (Applied Biosystems, California, USA). The sequences of the different primers provided by Applied Biosystems (California, USA) are presented in Table 1. Three PCR were performed: two multiplex for blaTEM, blaSHV, blaCTX-M-G2 and blaCTX-M-G9 [38] and one simplex for blaCTX-M-G1 [39].
| Bla genes | Sequence (5’-3’) | Fragments (pb) | References |
|---|---|---|---|
| TEM | For : CATTTCCGTGTCGCCCTTATTC Rev : CGTTCATCCATAGTTGCCTGAC |
800 | [38] |
| SHV | For: AGCCGCTTGAGCAAATTAAAC Rev: ATCCCGCAGATAAATCACCAC |
713 | [38] |
| CTX-M-G1 | For: GTTACAATGTGTGAGAAGCAG Rev: CCGTTTCCGCTATTACAAAC |
1000 | [39] |
| CTX-M-G2 | For: CGTTAACGGCACGATGAC Rev: CGATATCGTTGGTGGTRCCA* |
404 | [38] |
| CTX-M-G9 | For: TCAAGCCTGCCGATCTGGT Rev: TGATTCTCGCCGCTGAG |
561 | [38] |
Table 1: Sequences of targeted bla genes.
The PCR final volume of 50 μl, consisted of 2 μl of DNA, 25 μl of 1X AmpliTaq Gold (used as Master Mix), 5 μl Enhancer; 2 μl of each primer at different concentration (0.2 to 0.4 pmol/μl) and sterile water (qs). The amplification programs were:
BlaTEM/SHV and blaCTX-M-G2/G9: initial denaturation 94°C for 10 min. 30 cycles of denaturation 94°C for 40 s, annealing 60°C for 40 s and elongation 72°C for 1 min with a final elongation step at 72°C for 7 min.
BlaCTX-M-G1: initial denaturation 96°C for 10 min. 35 cycles of denaturation 94°C for 1 min, annealing 50°C for 1 min and elongation 72°C for 1 min with a final elongation step at 72°C for 10 min.
An Electrophoretic migration at 100 volts for 1 hour was performed on a 2% agarose gel stained with ethidium bromide to separate PCR products. A marker of 100 bp was used as reference. After migration, the various bands were observed under UV illumination and pictures recorded.
Statistical analysis
Statistical analyses were performed with the software Epi Info Version 7.1.1.14. Fisher's exact test with a significance level of 5% (p<0.05) which was used to interpret results.
Ethical consideration
This study received the INH approval for the transfer of Klebsiella spp. strains, to the molecular biology laboratory of CERBA/ LABIOGENE, University Ouaga I, Professor Joseph Ki-Zerbo, Burkina Faso. The institutional ethic committee of CERBA/LABIOGENE reviewed and approved the study protocol.
Results
Characteristics of Klebsiella spp. strains
A total of 64 strains of Klebsiella spp . resistant to at least one 3rd generation cephalosporin (ceftazidime, ceftriaxone, cefotaxime) or aztreonam were collected during the study period. They were isolated from urine 51.56% (33/64), vaginal swabs 32.81% (21/64), pus 12.5% (8/64) and sperm samples 3.13% (2/64). They included Klebsiella pneumoniae 85.94% (55/64) and Klebsiella oxytoca 14.06% (9/64).
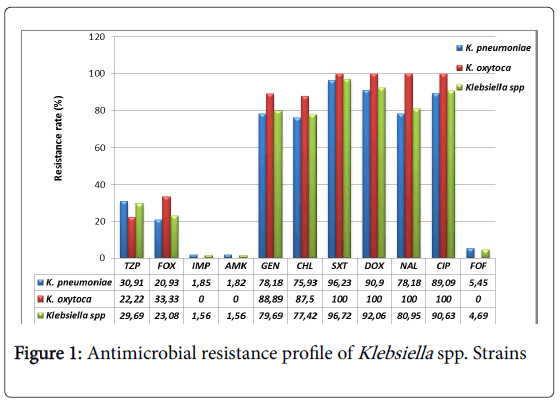
All Klebsiella pneumoniae strains were resistant to ceftazidime, cefepime and aztreonam. The resistance to ceftriaxone and cefotaxime was 98.18% (54/55). All (100%) Klebsiella oxytoca strains were resistant to ceftriaxone and cefepime and 88.89% (8/9) were resistant to ceftazidime, cefotaxime and aztreonam. The resistance profile to other β-lactams and other antibiotics is presented in Figure 1. Only imipenem, amikacin and fosfomycin are very active on Klebsiella spp. strains with a low resistance rate (below 5%). Ciprofloxacin, doxycycline and trimethoprim-sulfamethoxazole are the most inactive antibiotics with at least a resistance rate of 90%.
ESBL genes distribution within Klebsiella spp. strains
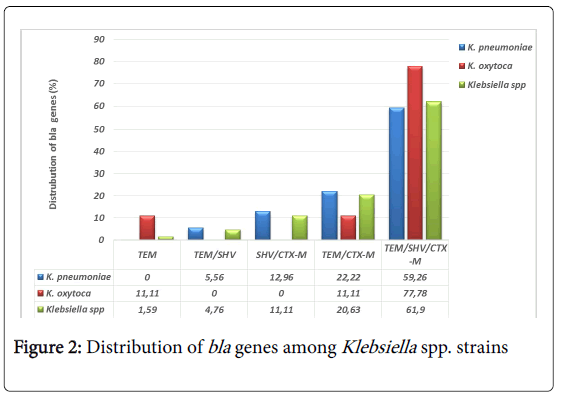
With regards to the PCR results, blaTEM, blaSHV and blaCTX-M group 1 (including blaCTX-M1, blaCTX-M3 and blaCTX-M15) were found in 98.44% (63/64) Klebsiella spp. strains. Of these, 98.41% (62/63) strains harbored a combination of blaTEM, blaSHV and blaCTX-M with predominance for the triple combination of TEM/SHV/CTX-M estimated at 61.90% (39/63) (Figure 2). Only one strain of Klebsiella oxytoca carried TEM gene. This strain was negative to the double disk test synergy for the phenotypically detection of ESBL. We did not identify any gene in one Klebsiella pneumoniae strain that did not show any ESBL phenotype.
Resistance to antibiotics according to the distribution of ESBL genes
Antibiotic resistance rate according to the distribution of genes in Klebsiella pneumoniae and Klebsiella oxytoca strains are summarized in Table 2. Results show that 100% Klebsiella pneumoniae strains (3/3) which expressed the first reported ESBL genes, TEM and SHV were resistant to ciprofloxacin and trimethoprim-sulfamethoxazole, 66.67% (2/3) were also resistant to piperacillin-tazobactam, gentamicin, chloramphenicol and nalidixic acid. All Klebsiella pneumoniae strains (7/7) carrying SHV and CTX-M genes were resistant to chloramphenicol and trimethoprim-sulfamethoxazole. Six (85.71%) of them were resistant to ciprofloxacin and gentamicin, 5 (71.43%) of them to doxycycline and nalidixic acid. All Klebsiella pneumoniae strains (12/12) carrying TEM and CTX-M genes were resistant to ciprofloxacin, doxycycline and trimethoprim-sulfamethoxazole while 90.91% (10/11) were resistant to chloramphenicol, 75% (9/12) to gentamicin and 66.67% (8/12) to nalidixic acid. Strikingly, the majority of studied strains, 32 Klebsiella pneumoniae and 7 Klebsiella oxytoca expressed simultaneously the three genes TEM, SHV and CTX-M. Among Klebsiella pneumoniae strains, 96.88% (31/32) were resistant to doxycycline, 96.67% (29/30) to trimethoprim-sulfamethoxazole, 87.5% (28/32) to nalidixic acid and ciprofloxacin, 81.25% (26/32) to gentamicin and 65.63% (21/32) to chloramphenicol. All Klebsiella oxytoca were resistant to gentamicin, nalidixic acid, ciprofloxacin, doxycycline and trimethoprim-sulfamethoxazole, while 83.33% (5/6) were resistant to chloramphenicol. Imipenem, amikacin and fosfomycin remained the most active antibiotics according to the ESBL genes distribution whereas one third (1/3) of strains expressing TEM and SHV genes are resistant to imipenem. The resistance rate to piperacillin-tazobactam ranged from 28.13% for the triple combination TEM/SHV/CTX-M to 66.67% for the double combination TEM/SHV in Klebsiella pneumoniae. Similarly, the resistance rate to cefoxitin ranged from 17.39% for the triple combination TEM/SHV/CTX-M to 33.33% for the double combination TEM/SHV. We did not find statistically significant differences for these two antibiotics (p=0.472 and p=0.426>0.05) between the groups expressing three ESBL genes (TEM/SHV/CTX-M) or two ESBL genes (TEM/SHV, SHV/CTX-M, TEM/CTX-M).
| ATB | TEM | TEM/SHV | SHV/CTX-M | TEM/CTX-M | TEM/SHV/CTX-M | ||
|---|---|---|---|---|---|---|---|
| Ko# (n/N) | K (%, n/N) | Kp (%, n/N) | Kp (%,n/N) | Ko# (n/N) | Kp (%,n/N) | Ko (%,n/N) | |
| TZP | 0/1 | 66.67 2/3 | 14.29 1/7 | 41.67 5/12 | 0/1 | 28.13 9/32 | 28.57 2/7 |
| FOX | 1/1 | 33.33 1/3 | 14.29 1/7 | 22.22 2/9 |
0/1 | 17.39 4/23 | 28.57 2/7 |
| IPM | 0/1 | 33.33 1/3 | 0 0/7 | 0 0/12 |
0/1 | 0 0/32 | 0 0/7 |
| AMK | 0/1 | 0 0/3 | 0 0/7 | 0 0/12 |
0/1 | 3.13 1/32 | 0 0/7 |
| GEN | 0/1 | 66.67 2/3 | 85.71 6/7 | 75 9/12 |
1/1 | 81.25 26/32 | 100 7/7 |
| CHL | 1/1 | 66.67 2/3 | 100 7/7 | 90.91 10/11 | 1/1 | 65.63 21/32 | 83.33 5/6 |
| FOF | 0/1 | 0 0/3 | 0 0/7 | 8.33 1/12 |
0/1 | 3.13 1/32 | 0 0/7 |
| NAL | 1/1 | 66.67 2/3 | 71.4 5/7 | 66.67 8/12 | 1/1 | 87.5 28/32 | 100 6/6 |
| CIP | 1/1 | 100 3/3 | 85.71 6/7 | 100 12/12 |
1/1 | 87.5 28/32 | 100 7/7 |
| DOX | 1/1 | 66.67 2/3 | 71.43 5/7 | 100 12/12 |
1/1 | 96.88 31/32 | 100 6/6 |
| SXT | 1/1 | 100 3/3 | 100 7/7 | 100 12/12 |
1/1 | 96.67 29/30 | 100 6/6 |
Table 2: Antimicrobial resistance profile of Klebsiella spp. strains according to the distribution of bla genes. ATB: Antibiotic, Kp: Klebsiella pneumoniae, Ko: Klebsiella oxytoca, TZP: piperacillintazobactam, FOX; cefoxitin, IPM: imipenem, AMK: amikacin, GEN: gentamicin, CHL: chloramphenicol, FOF: fosfomycin, NAL: nalidixic acid, CIP: ciprofloxacine, DOX: doxycycline, SXT: trimetroprimsulfamethoxazole; *The non-ESBL strain, Klebsiella pneumoniae was resistant to ceftriaxone, ceftazidime, cefotaxime, cefepime and aztreonam. The strain was also negative to the double disk synergy test for ESBL detection. #The proportions was not indicated for the effect if of one strain.
Distribution of the combination TEM/SHV/CTX-M according to the biological specimen
The combination of TEM, SHV and CTX-M genes was carried by 57.14% (12/21), 45.45% (15/33) and 37.5% (3/8) of Klebsiella spp. strains isolated from vaginal swabs, urine and pus respectively. Klebsiella pneumoniae from the two sperm samples expressed simultaneously these three genes, TEM, SHV and CTX-M.
Discussion
Nowadays, resistance to antibiotics is a real public health concern worldwide. It has been clearly demonstrated that the production of β- lactamases is the most important mechanism of antibiotic resistance in Gram negative bacteria. The description of first β-lactamases TEM-1/2, SHV-1 and their derivatives, was followed by the characterization of new ones dominated by the CTX-M group which is now, the most widespread. This study was carried out to characterize ESBL genes in Klebsiella spp. strains resistant to at least one 3rd generation cephalosporins or aztreonam, isolated from different specimen received at INH Lomé, Togo. It has been reported that Klebsiella spp. present a high medical interest among other Enterobacteriaceae as expressing ESBL resistance mechanism and are frequently isolated in hospital and community infections.
According to our expectation, all Klebsiella spp. strains were highly resistant (over 98%) to 3rd generation cephalosporins (ceftazidime, ceftriaxone, cefotaxime), 4th generation (cefepime) and aztreonam. However, their resistance rate is higher than those found in previous studies conducted in North America. Indeed, through the international monitoring program for antibiotic resistance SENTRY involving four geographic regions, Asia Pacific, Europe, Latin America, North America (United States of America and Canada), Sader et al. found a resistance rate of 80.3% for ceftazidime, 62.1% for ceftriaxone, 83.9% for aztreonam and 10.6% for cefepime [40]. Nevertheless, our results are in line with those of Irenge et al. showing a 100% prevalence of resistance for ceftriaxone and ceftazidime on ESBL producing Klebsiella spp. strains isolated from urine in Congo [41]. For piperacillin-tazobactam and cefoxitin two β-lactams subject of comparative study on their efficiency on ESBL producing Enterobacteriaceae [42-44], we found a prevalence of resistance at 30.91% and 20.93% for Klebsiella pneumoniae and 22.22%, 33.33% for Klebsiella oxytoca respectively. These prevalence rates seem low compared to those reported in North America, 47.4% for piperacillintazobactam, and 44.2% for cefoxitin [40]. The resistance to cefoxitin can be induced by a production of cephalosporinase AmpC, a class A enzyme that inactivates other 3rd generation cephalosporins. AmpC decreases the intracellular concentration of the antibiotic by the efflux pump mechanism and mediates a loss of outer membrane porins [45]. Fortunately, Klebsiella spp. isolated in this study have remained relatively sensitive to imipenem, amikacin and fosfomycin which are among other, recommended antibiotics in case of infection by ESBL producing bacteria [46-48]. The prevalence of resistance to these antibiotics ranged from 0 to 5% as in a series of ESBL producing Klebsiella spp. strains isolated in Tunisia, where the prevalence of resistance to imipenem was nulle [49]. However, the resistance rates for amikacin and fosfomycin were high, 10% and 17.5%, respectively [49]. Higher resistance rates were found in non-intensive care units in the United States of America and Europe for imipenem (22% and 6.9%) and amikacin (18.9% and 19.1%), respectively [50].
In addition, Klebsiella spp. isolated in the present study showed a strong resistance rate to quinolones (nalidixic acid and ciprofloxacin) ranging from 78.18% (43/55) 89.09% (49/55) for Klebsiella pneumoniae to 100% for Klebsiella oxytoca. Similar results were found in Congo, in Tunisia and in Bolivia. In Congo, 83.3% of Klebsiella spp. strains were resistant to ciprofloxacin [41], and in Tunisia, 80% (32/40) of Klebsiella spp. strains were resistant to nalidixic acid and 67.5% (27/40) to ciprofloxacin [49]. Results in Bolivia showed that 100% (6/6) of Klebsiella spp. strains were resistant to ciprofloxacin and nalidixic acid [51].
The analysis of the PCR product showed the simultaneous presence of the three genes, TEM, SHV and CTX-M in 61.90% of the studied strains, 59.26% (32/54) in Klebsiella pneumoniae, 77.78% (7/9) in Klebsiella oxytoca. TEM was detected in 1.59% (1/9) Klebsiella oxytoca strain, SHV gene alone was not found in this study. In Burkina Faso, on 28 ESBL producing Klebsiella pneumoniae screened for the detection of bla genes, 14.28% expressed TEM, 10.71% SHV, 25% CTX-M, 10.71% TEM and SHV and 3.57% of strains exhibited the triple combination of TEM, SHV and CTX-M [52]. In another study conducted in Burkina Faso, CTX-M15 was found in 17/17 Klebsiella spp. isolated from urine in a pediatric hospital in Ouagadougou [53] suggesting that CTX-M15 was present in West Africa. Our results are consistent with those found by Alibi et al. in Tunisia, on a large series of 118 ESBL producing Klebsiella spp. where the triple combination TEM/SHV/CTX-M represented 44.91% (53/118), and the double combinations were SHV/CTX-M represented 28.81% (34/118) of cases, TEM/CTX-M represented 4.23% (5/118), and TEM/SHV represented 3.39% (4/118) of cases [54]. The presence of TEM, SHV and CTX-M genes alone in the study by Alibi et al. in Tunisia was high compared to our results with 3.39% (4/118), 4.23% (5/118) and 11% (13/118), respectively [54]. In Mozambique, on 19 ESBL producing Klebsiella pneumoniae isolated from urine and blood culture, Pons et al., found 15.79% (3/19) expressed only CTX-M15, 63.16% (12/19) TEM/CTX-M15 and 21.05% (4/19) the triple combination TEM/SHV/ CTX-M15 [55]. In Tanzania, on 92 ESBL producing Klebsiella pneumoniae, Mshana et al. found 53.26% (49/92) carried TEM/CTXM15, 11.96% (11/92) SHV/CTX-M15 and 10.87% (10/92) CTX-M15 alone [56]. Similarly, in Brazil, Jaskulski et al. found that 16.67% (2/12) of ESBL producing Klebsiella pneumoniae carried TEM, CTX-M, TEM/SHV genes, while 8.33% (8/12) carried SHV alone. The double TEM/CTX-M and triple TEM/SHV/CTX-M combinations were 25% (3/12) and 16.67% (2/12) respectively [57]. It is observed that the expression of blaCTX-M is currently more prevalent and as we report for the first time in Togo, more frequent in a triple combination with blaTEM and blaSHV. The explanation of this triple combination should be more investigated.
The antimicrobial susceptibility profile of ESBL producing Klebsiella spp. strains shows that the different ESBL genes do not significantly influence the activity of the antibiotics. As expected, resistance to imipenem, amikacin and fosfomycin was low. On the contrary, the resistance rates to trimethoprim-sulfamethoxazole, ciprofloxacin, nalidixic acid, chloramphenicol and gentamicin were very high ranging from 65.63% for chloramphenicol in the triple combination TEM/SHV/CTX-M to 100% for most of them in all double combinations. These antimicrobial resistances associated with ESBL genes are in line with our expectation and are consistent with the results found in previous studies [1–6]. Indeed, it has been described that ESBL genes and some antibiotic resistance genes are often found on the same mobile genetic elements and are thereby transmitted together by horizontal transfer [1–6]. In Tanzania, on ESBL producing Klebsiella pneumoniae causing sepsis in neonates, Mshana et al. showed that blaCTX-M15 was localized on a plasmid of 25 to 485KB and that ST14 and ST48 clones carried also resistance genes to gentamicin and trimethoprim-sulfamethoxazole [56]. More recently, in a study in Central African Republic, Rafai et al. showed that all ESBL producing Klebsiella pneumoniae isolated from surgical wounds, expressed blaCTX-M15 and aac(6 ')-Ib-cr [58]. The last one has been described to be associated with a resistance to ciprofloxacin, norfloxacin and aminoglycosides [59]. This is probably due to the same mechanisms that might explain the results we observed.
In this study, the combination of TEM, SHV and CTX-M genes was found on strains isolated mostly from urine and sperm. This result is also consistent with previous findings in Tunisia where 86.44% of ESBL producing Klebsiella spp. were isolated from urine and 44.91% (53/118) of all studied strains expressed the triple combination TEM/SHV/CTX-M [54]. The observation of this combination based on the pathological sample needs to be analyzed more thoroughly.
Conclusion
In this study, we observed that the majority of Klebsiella spp. resistant to 3rd generation cephalosporins were producing ESBL. The major groups of ESBL genes, TEM, SHV and CTX-M were present. We report for the first time, that the triple combination of TEM, SHV and CTX-M genes was predominant. Stressfully, all ESBL producing Klebsiella oxytoca were resistant to quinolones (nalidixic acid and ciprofloxacin), doxycycline and trimethoprim-sulfamethoxazole and also to a lesser extent to gentamicin and chloramphenicol. The same trend is observed for ESBL producing Klebsiella pneumoniae . Antibiotics that remained active, including fosfomycin, amikacin and imipenem are unfortunately a little affordable to patients in limited resources countries. It is therefore important to these countries to strengthen public health policies in order to prevent, monitor and control antimicrobial resistance. In addition, some work has been done for screening of medicinal plants with potential antimicrobial activities. In a public health point of view, it is now important for political, economic and medical sectors to work together towards the selection of those plants which could be active against clinical resistant strains.
Author's Contribution
SD, FS, SK and JS designed the study, FS, SS, AO, AS, AK, TZ and AD performed the experiments, FS, CN and SD analyzed the data and wrote the manuscript which was approved by all the other co-authors.
Funding Information
This study was conducted through the scholarship granted by the Islamic Development Bank for an enrolment in the Master II of molecular biology and genetics at the University of Ouaga I Prof. Joseph Ki-Zerbo, Burkina Faso. The WAEMU through the PACER II program granted the molecular biology laboratory CERBA/ LABIOGENE University Ouaga I Prof. Joseph Ki-Zerbo for the realization of molecular biology analysis. The funders had no role in the writing up of this paper.
Acknowledgments
We are grateful to the National Institute of Health (INH) Lomé, Togo for the collection of Klebsiella spp. strains and the antimicrobial susceptibility testing. We thank Aristide Bado and Yacinthe Raogo Zabré for statistical guidance. The authors thank the funders for financial support.
References
- Bush K, Jacoby GA (2010) Updated Functional Classification of β-Lactamases. AntimicrobAgents Chemother 54: 969-976.
- Drawz SM, Bonomo RA (2010) Three Decades of β-Lactamase Inhibitors. ClinMicrobiolRev 23: 160-201.
- Philippon A, Labia R, Jacoby G (1989) Extended-spectrum beta-lactamases. Antimicrob Agents Chemother33: 1131.
- Jacoby GA, Medeiros AA (1991) More extended-spectrum beta-lactamases. Antimicrob Agents Chemother35: 1697.
- Bradford PA (2001) Extended-Spectrum β-Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of This Important Resistance Threat. ClinMicrobiol Rev 14: 933-951.
- Paterson DL, Bonomo RA (2005) Extended-Spectrum β-Lactamases: a Clinical Update. ClinMicrobiol Rev 18: 657-686.
- Woodford N (2004) Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum β-lactamases in the UK. J AntimicrobChemother54: 735-743.
- Lewis JS, Herrera M, Wickes B, Patterson JE, Jorgensen JH (2007) First Report of the Emergence of CTX-M-Type Extended-Spectrum β-Lactamases (ESBLs) as the Predominant ESBL Isolated in a U.S. Health Care System. Antimicrob Agents Chemother51: 4015-4021.
- Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, et al. (2007) CTX-M: changing the face of ESBLs in Europe. J AntimicrobChemother59: 165-174.
- Cantón R, Novais A, Valverde A, Machado E, Peixe L, et al. (2008) Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Europe. ClinMicrobiol Infect 14: 144-153.
- Rossolini GM, D’andrea MM, Mugnaioli C (2008) The spread of CTX-M-type extended-spectrum β-lactamases. ClinMicrobiol Infect 14: 33-41.
- Arpin C, Quentin C, Grobost F, Cambau E, Robert J, et al. (2009) Nationwide survey of extended-spectrum β-lactamase-producing Enterobacteriaceae in the French community setting. J AntimicrobChemother63: 1205-1214.
- Woerther P-L, Burdet C, Chachaty E, Andremont A (2013) Trends in Human Fecal Carriage of Extended-Spectrum β-Lactamases in the Community: Toward the Globalization of CTX-M. ClinMicrobiol Rev26: 744-758.
- Zhang J, Zheng B, Zhao L, Wei Z, Ji J, et al. (2014) Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect Dis 14: 659.
- Ahmed SH, Daef EA, Badary MS, Mahmoud MA, Abd-Elsayed AA (2009) Nosocomial blood stream infection in intensive care units at Assiut University Hospitals (Upper Egypt) with special reference to extended spectrum β-lactamase producing organisms. BMC Res Notes 2: 76.
- Jean S-S, Hsueh PR (2011) High burden of antimicrobial resistance in Asia. Int J AntimicrobAgents37: 291-295.
- Peralta G, Lamelo M, Álvarez-García P, Velasco M, Delgado A, et al. (2012) Impact of empirical treatment in extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella spp. bacteremia. A multicentric cohort study. BMC Infect Dis 12: 1.
- Christopher A, Mshana SE, Kidenya BR, Hokororo A, Morona D (2013)Bacteremia and resistant gram-negative pathogens among under-fives in Tanzania. Ital JPediatr39: 1.
- Pereira CAP, Marra AR, Camargo LFA, Pignatari ACC, Sukiennik T, et al. (2013) Nosocomial Bloodstream Infections in Brazilian Pediatric Patients: Microbiology, Epidemiology, and Clinical Features. Safdar N, editor. PLoS ONE8: e68144.
- Hawser SP, Bouchillon SK, Hoban DJ, Badal RE, Canton R (2010) Incidence and Antimicrobial Susceptibility of Escherichia coli and Klebsiellapneumoniae with Extended-Spectrum β-Lactamases in Community and Hospital-Associated Intra-Abdominal Infections in Europe: Results of the 2008 Study for Monitoring Antimicrobial Resistance Trends (SMART). Antimicrob Agents Chemother54: 3043-3046.
- Fernández-Canigia L, Dowzicky MJ (2012) Susceptibility of important Gram-negative pathogens to tigecycline and other antibiotics in Latin America between 2004 and 2010. Ann ClinMicrobiolAntimicrob11: 1-9.
- Lu PL, Liu YC, Toh HS, Lee YL, Liu YM, et al. (2012) Epidemiology and antimicrobial susceptibility profiles of Gram-negative bacteria causing urinary tract infections in the Asia-Pacific region: 2009–2010 results from the Study for Monitoring Antimicrobial Resistance Trends (SMART). Int J Antimicrob Agents 40: S37-S43.
- Podschun R, Ullmann U (1998) Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. ClinMicrobiol Rev 11: 589-603.
- Brisse S, Grimont F, Grimont PAD (2006) The Genus Klebsiella. The Prokaryotes 3: 159-96.
- Blomberg B, Jureen R, Manji KP, Tamim BS, Mwakagile DSM, et al. (2005) High Rate of Fatal Cases of PediatricSepticemia Caused by Gram-Negative Bacteria with Extended-Spectrum Beta-Lactamases in Dar es Salaam, Tanzania. J ClinMicrobiol43: 745-749.
- Tuon FF, Kruger M, Terreri M, Penteado-Filho SR, Gortz L (2011)Klebsiella ESBL bacteremia-mortality and risk factors. Braz J Infect Dis15: 594-598.
- Girometti N, Lewis RE, Giannella M, Ambretti S, Bartoletti M, et al. (2014)Klebsiellapneumoniae Bloodstream Infection: Epidemiology and Impact of Inappropriate Empirical Therapy. Medicine (Baltimore) 93: 298-309.
- Akoachere JFTK, Yvonne S, Akum NH, Seraphine EN (2012) Etiologic profile and antimicrobial susceptibility of community-acquired urinary tract infection in two Cameroonian towns. BMC Res Notes 5: 1.
- Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, et al. (2015) Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiellapneumoniae , an urgent threat to public health. ProcNatlAcadSci 112: E3574-381.
- Okeke IN, Lamikanra A, Edelman R (1999) Socioeconomic and behavioral factors leading to acquired bacterial resistance to antibiotics in developing countries. Emerg Infect Dis5: 18-27.
- Monnet DL (2000) Consommationd’antibiotiques et résistance bactérienne. Ann FrAnesthRéanimation [Internet] Elsevier. 409-417.
- Peleg AY, Hooper DC (2010) Hospital-Acquired Infections Due to Gram-Negative Bacteria. N Engl J Med 362: 1804-1813.
- Bertrand X, Dowzicky MJ (2012) Antimicrobial Susceptibility Among Gram-Negative Isolates Collected From Intensive Care Units in North America, Europe, the Asia-Pacific Rim, Latin America, the Middle East, and Africa Between 2004 and 2009 as Part of the Tigecycline Evaluation and Surveillance Trial. ClinTher34: 124-137.
- Hoban DJ, Lascols C, Nicolle LE, Badal R, Bouchillon S, et al. (2010) Antimicrobial susceptibility of Enterobacteriaceae, including molecular characterization of extended-spectrum beta-lactamase–producing species, in urinary tract isolates from hospitalized patients in North America and Europe: results from the SMART study 2009–2010. DiagnMicrobiol Infect Dis74: 62-67.
- Salou M, Assimadzi K, Wateba IM, Dossim S, Tigossou SD, et al. (2011) Resistance aux antibiotiques des bactériesisolées en 2009 au laboratoire de bacteriologie du Chu-TokoinLome-Togo. J RechSciUnivLome 13: 151-159.
- Bonnet R, Caron F, Cavallo J, Chardon H, Chidiac C, et al. (2013) Comité de l’antibiogramme de la sociétéfrançaise de microbiologie. Recommandations [Internet] 19: 133-142
- Jarlier V, Nicolas MH, Fournier G, Philippon A (1988) Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis 10: 867-878.
- Dallenne C, Da Costa A, Decre D, Favier C, Arlet G (2010) Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J AntimicrobChemother65: 490-495.
- Pagani L, Dell’Amico E, Migliavacca R, D’Andrea MM, Giacobone E, et al. (2003) Multiple CTX-M-Type Extended-Spectrum β-Lactamases in Nosocomial Isolates of Enterobacteriaceae from a Hospital in Northern Italy. J ClinMicrobiol41: 4264-4269.
- Sader HS, Hsiung A, Fritsche TR, Jones RN (2007) Comparative activities of cefepime and piperacillin/tazobactam tested against a global collection of Escherichia coli and Klebsiella spp. with an ESBL phenotype.DiagnMicrobiol Infect Dis57: 341-344.
- Irenge LM, Kabego L, Vandenberg O, Chirimwami RB, Gala JL (2014) Antimicrobial resistance in urinary isolates from inpatients and outpatients at a tertiary care hospital in South-Kivu Province (Democratic Republic of Congo). BMC Res Notes 7: 374.
- Babini GS, Yuan M, Hall LMC, Livermore DM (2003) Variable susceptibility to piperacillin/tazobactam amongst Klebsiella spp. with extended-spectrum β-lactamases. J AntimicrobChemother51: 605-612.
- Guet-Revillet H, Emirian A, Groh M, Nebbad-Lechani B, Weiss E, et al. (2014) Pharmacological Study of Cefoxitin as an Alternative Antibiotic Therapy to Carbapenems in Treatment of Urinary Tract Infections Due to Extended-Spectrum-β-Lactamase-Producing Escherichia coli. Antimicrob Agents Chemother58: 4899-4901.
- Tamma PD, Han JH, Rock C, Harris AD, Lautenbach E, et al. (2015)Carbapenem Therapy Is Associated With Improved Survival Compared With Piperacillin-Tazobactam for Patients With Extended-Spectrum β-Lactamase Bacteremia. Clin Infect Dis Off Publ Infect Dis Soc Am 60: 1319-1325.
- Jacoby GA (2009) AmpC-Lactamases. ClinMicrobiol Rev22: 161-182.
- Paterson DL (2000) Recommendation for treatment of severe infections caused by Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs). ClinMicrobiol Infect6: 460-463.
- Gupta K, Hooton TM, Naber KG, Wullt B, Colgan R, et al. (2011) International Clinical Practice Guidelines for the Treatment of Acute Uncomplicated Cystitis and Pyelonephritis in Women: A 2010 Update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis52: e103-120.
- Barber AE, Norton JP, Spivak AM, Mulvey MA (2013) Urinary Tract Infections: Current and Emerging Management Strategies. Clin Infect Dis 57: 719-724.
- Khalifa ABH, Khedher M (2012)Epidémiologie des souches de Klebsiella spp. uropathogènesproductrices de β-lactamases à spectre élargidans un hôpitaluniversitaireTunisien, 2009. PatholBiol60: e1-e5.
- Sader HS, Farrell DJ, Flamm RK, Jones RN (2014) Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalized in intensive care units in United States and European hospitals (2009–2011). DiagnMicrobiol Infect Dis78: 443-448.
- Bartoloni A, Sennati S, Di Maggio T, Mantella A, Riccobono E, et al. (2016) Antimicrobial susceptibility and emerging resistance determinants (blaCTX-M, rmtB, fosA3) in clinical isolates from urinary tract infections in the Bolivian Chaco. Int J Infect Dis43: 1-6.
- Zongo K, MetuorDabiré A, Compaoré LG, Sanou I, Sangaré L, et al. (2015) First detection of bla TEM, SHV and CTX-M among Gram negative bacilli exhibiting extended spectrum β-lactamase phenotype isolated at University Hospital Center, YalgadoOuedraogo, Ouagadougou, Burkina Faso. Afr JBiotechnol14: 1174-1180.
- MetuorDabiré A, Zongo KJ, Zéba B, Moussawi J, Boucher M, et al. (2013) Resistances to the Oxyimino-Cephalosporins by Ctx-M-15 Producing Klebsiella Isolated From the Urines Samples of Patients in the University Hospital Complex Paediatric Charles De Gaulle (Chup-Cdg) Of Ouagadougou in Burkina Faso. J Asian SciRes 3: 882.
- Alibi S, Ferjani A, Boukadida J (2015) Molecular characterization of extended spectrum beta-lactamases produced by Klebsiellapneumoniae clinical strains from a Tunisian Hospital. Médecine MalInfect45: 139-143.
- Pons MJ, Vubil D, Guiral E, Jaintilal D, Fraile O, et al. (2015) Characterisation of extended-spectrum β-lactamases among Klebsiellapneumoniae isolates causing bacteraemia and urinary tract infection in Mozambique. J GlobAntimicrob Resist3: 19-25.
- Mshana SE, Hain T, Domann E, Lyamuya EF, Chakraborty T, et al. (2013) Predominance of Klebsiellapneumoniae ST14 carrying CTX-M-15 causing neonatal sepsis in Tanzania. BMC Infect Dis 13: 466.
- Rafaï C, Frank T, Manirakiza A, Gaudeuille A, Mbecko JR, et al. (2015) Dissemination of IncF-type plasmids in multiresistant CTX-M-15-producing Enterobacteriaceae isolates from surgical-site infections in Bangui, Central African Republic. BMC Microbiol 15: 15.
- Jaskulski MR, Medeiros BC, Borges JV, Zalewsky R, Fonseca MEC, et al. (2013) Assessment of extended-spectrum β-lactamase, KPC carbapenemase and porin resistance mechanisms in clinical samples of Klebsiellapneumoniae and Enterobacter spp. Int J Antimicrob Agents42: 76-79.
- Jacoby GA (2005) Mechanisms of Resistance to Quinolones. Clin Infect Dis 41: S120-126.
Relevant Topics
- Back Pain Diagnosis
- Cardiovascular Diagnosis
- Clinical Diagnosis
- Clinical Echocardiography
- COPD Diagnosis
- Diabetes Diagnosis
- Diagnosis Methods
- Diagnosis of cancer
- Diagnosis of CNS
- Diagnosis of Diabetes
- Diagnostic Products
- Diagnostics Market Analysis
- Heart diagnosis
- Immuno Diagnosis
- Infertility Diagnosis
- Medical Diagnostic Tools
- Preimplementation Genetic Diagnosis
- Prenatal Diagnostics
- Ultrasonography
Recommended Journals
Article Tools
Article Usage
- Total views: 7683
- [From(publication date):
December-2016 - Apr 02, 2025] - Breakdown by view type
- HTML page views : 6484
- PDF downloads : 1199