Detecting and Isolating False Negatives of SARS-Cov-2 Primers and Probe Sets among the Japanese Population: A Laboratory Testing Methodology and Study
Received: 12-Feb-2021 / Accepted Date: 26-Feb-2021 / Published Date: 05-Mar-2021 DOI: 10.4172/2165-7386.s1.10004
Abstract
Objective: The study was performed for a comparative analysis between primers from Japan’s and US’s disease control centers as well as to investigate the virus sequence alignment with primers’ oligonucleotide to introduce primer sets of high detectability with reduce false negative results.
Design or methods: 11,652 samples from Japanese population were tested for Novel Severe Acute Respiratory Syndrome Corona Virus-2 (SARS-CoV-2) positive using recommended RT-PCR primer-probe sets from Japan National Institute of Infectious Disease (NIID) and US Centers for Disease Control and Prevention (CDC). Primer-probe sensitivity was analyzed for higher detectability for SARSCoV- 2 positive cases.
Results: Of the 102 positive samples, 17 samples (16.7% of total positives) showed inconsistent results when tested simultaneously for the following primers: JPN-N2, JPN-N1, CDC-N1, and CDC-N2. Our results revealed that CDC recommended primer-probe sets showed relatively higher detection sensitivity and accuracy. Further, virus sequence alignment analysis showed evidences for virus mutation occurred at primer’s binding sites.
Conclusion: The inconsistency in the RT-PCR results for SARS-CoV-2 detection using JPN-N1, JPN-N2, CDC-N1, and CDC-N2 primer-probe sets could be attributed to differences in primer efficiency or/and virus mutation at primer-probe’ binding sites. The use of JPN-N2 combined with CDC-N2 and CDC-N1 primer-probe sets produce the most effective results as well as may reduce the false negatives results in Japan and overseas.
Keywords: COVID-19; SARS-CoV-2; RT-PCR performance; Genomic variants; Primers; Sensitivity
Introduction
The global COVID-19 pandemic has spread across various continents in diverse methods and speed while opening up discussion for technological and scientific questions pertaining methodology of testing accuracy among diverse viral strains.
On the issue of testing sensitivity and accuracy, RT-PCR has been the gold standard testing method for SARS-CoV-2 detection compare to other rapid testing methods [1,2]. However, each country’s infectious disease authority has established its own guidelines, primerprobe sets, and protocols, thereby; the global testing community lacks a “Standardized Universal Primer(s)” (SUP) that is foolproof for the COVID-19 patients among various populations today. As a result, RTPCR testing accuracy and results may vary depending on which primer was used. Low primer efficiency or/and mutation at the primer-probe binding sites as well as very low viral RNA in the sample may resulting in false negative RT-PCR results.
There are two commonly known factors associated with inaccurate testing results for COVID-19 using RT-PCR testing method. First is the failure to retrieve sufficient amounts of viral RNA present in the sample is administered or the method of how a sample is collected. For example, a nasopharyngeal swab may be unable to obtain sufficient amounts of RNA if it does not come in contact at a nasal position where the presence of the virus is concentrated. In the case of saliva collection kit, use during the first several days of viral contact may result in insufficient amounts of RNA.
For both nasopharyngeal swab and saliva kit, low amounts of viral load occurring at later stage among discharged or recovering patients has tendency to show extremely low RNA count. While many countries have issued a standard 14-day quarantine, Genesis Healthcare, a licensed clinical laboratory in Tokyo, Japan, has confirmed through tested samples where the recovering and discharged patient is still testing positive after 14 day despite low RNA count. Though this confirmation does not necessarily imply that the patient is infectious after 14 days of quarantine as observed in a study [3], while the test result remains positive, it supports recent study which outlines the long duration of the RNA-positive tail and calls for reconsideration of containment strategy [4].
The second factor is the lack of a standardized international testing method for COVID-19 using a set of common primer(s) among different nations/populations or hereby referred to as SUP. Different primers used by different country’s testing protocols prevent effective tracking of pandemic due to differences in false negative results due to viral genetic variation which may have been introduced from different regions.
Researchers have started to recognize this problem as more SARS-CoV- 2 genomic data are becoming available. In particular, a researcher group evaluated five assay panels for possible loss of detection sensitivity due to genetic variability of the virus [5] which was consistent with the data observed by Genesis Healthcare.
The current study of summarizing the RT-PCR result discrepancy for SARS-CoV-2 testing among different primers recommended by NIID and CDC will be of great interest to the scientific community working on infectious diseases detection, diagnosis methods and health care.
Methods
Study design
Japan’s COVID-19 RT-PCR public health observation is unique in that while Wuhan, China was locked down from January 23, 2020 till April 8th, 2020; Japan’s border was still open to remainder of China until March 9th, 2020 and to US and Europe until March 26, 2020. This time gap in closing the international border leaves an inquiry that COVID-19 could have possibly entered Japan at different times from both East and West.
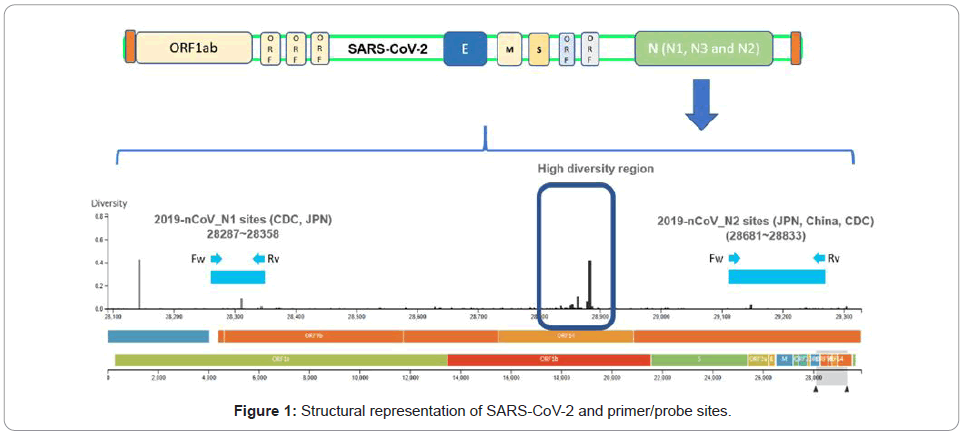
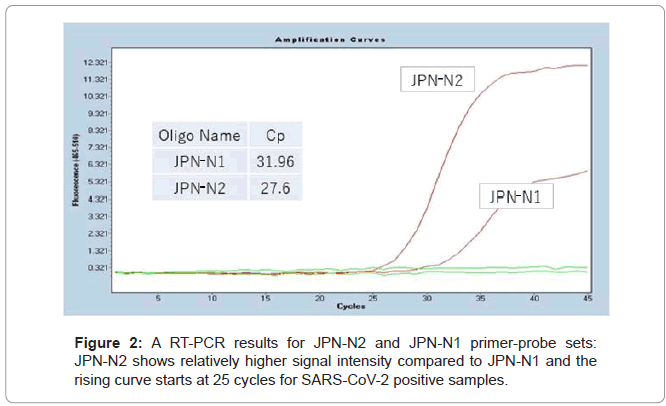
On March 11th and March 19th, NIID announced the testing protocols for COVID-19 testing by outlining two primer-probe sets JPN-N2 and JPN-N1 targeting the nucleocapsid region, N of SARS-CoV- 2 as shown in (Figures 1 and 2). The diversity sites were sourced from [6].
JPN-N2 showed consistently higher primer sensitivity than JPN-N1. As a result, it is sought that NIID later eliminated JPN-N1 to only reflect JPN-N2 in calling positive samples.
Due to the continuing dialogue by the medical community to retest samples whose results were not considered to reflect patient’s symptoms, testing JPN-N1 was continued and CDC-N1 and CDC-N2, both of which are located in nucleocapsid region N of SARS-CoV-2, were added to identify whether JPN-N2 primer was adequate to identify positive cases for all samples.
Sample collection
All COVID-19 positive testing samples were obtained between April 2020 and August 2020 and were either obtained from saliva or nasopharyngeal test kits. The nasopharyngeal testing included two swabs per person and were administered to maximize virus collection to rule out miscalling due to low RNA count. Both nasopharyngeal and saliva samples were immediately immersed in RNA preservation solution after collection to inactivate the virus while maintaining RNA stability during transport. Samples were delivered to the Genesis Healthcare’s PCR testing facility within 24 hours at temperatures between 20°C and 27°C. RNA extraction was immediately conducted followed by RT-PCR test. The entire RNA extraction to RT-PCR testing process was completed within 6 hours of receipt at the same laboratory using the below testing method.
COVID-19 RNA collection method, extraction and RT-PCR testing method
Saliva samples were collected using saliva RNA sample collection kit by Zeesan Biotech Co., Ltd. Nasopharyngeal swab samples were collected using virus RNA sample collection kit by Zeesan Biotech Co., Ltd. To remove viscosity, 0.5 ml of 20% DTT was added to each saliva sample followed by, vortex, incubation at 50° Celsius for 10 minutes, and then centrifuged at 1,500 rpm for 10 minutes.
180 μl of supernatant of DTT-treated saliva samples and 180 μl of nasopharyngeal swab suspension were used for RNA extraction. RNA extraction was performed by MGISP-960 system (MGI Tech Co., Ltd). RT-qPCR for specific amplification of the N gene or was performed using Light Cycler 480 system Ⅱ (Roche Diagnostics K.K.). Final reaction volume was 12.5 μl, including 2.0 μl of RNA template, 6.25 μl of One Step Prime Script III RT-qPCR Mix (TAKARA BIO INC.), and forward primer, reverse primer and probe (primer and probe sequence, final concentration was shown in Table 1.
| Primer/ probe set | Primer name | Description | Oligonucleotide sequence (5’>3’) | Label | Working conc. |
|---|---|---|---|---|---|
| JPN-N1 | JPN-N1-F | N_Sarbeco_F1 | CACATTGGCACCCGCAATC | None | 0.48 mM |
| JPN-N1-R | N_Sarbeco_R1 | GAGGAACGAGAAGAGGCTTG | None | 0.64 mM | |
| JPN-N1-P | N_Sarbeco_P1 | ACTTCCTCAAGGAACAACATTGCCA | FAM, BHQ-1 | 0.16 mM | |
| JPN-N2 | JPN-N2-F | NIID_2019-nCOV_N_F2 | AAATTTTGGGGACCAGGAAC | None | 0.4 mM |
| JPN-N2-R | NIID_2019-nCOV_N_R2 | TGGCAGCTGTGTAGGTCAAC | None | 0.56 mM | |
| JPN-N2-P | NIID_2019-nCOV_N_P2 | ATGTCGCGCATTGGCATGGA | FAM, BHQ-1 | 0.16 mM | |
| CDC-N1 | CDC-N1-F | CDC_2019-nCoV_N1-F | GACCCCAAAATCAGCGAAAT | None | 0.5 mM |
| CDC-N1-R | CDC_2019-nCoV_N1-R | TCTGGTTACTGCCAGTTGAATCTG | None | 0.5 mM | |
| CDC-N1-P | CDC_2019-nCoV_N1-P | ACCCCGCATTACGTTTGGTGGACC | FAM, BHQ-1 | 0.2 mM | |
| CDC-N2 | CDC-N2-F | CDC_2019-nCoV_N2-F | TTACAAACATTGGCCGCAAA | None | 0.5 mM |
| CDC-N2-R | CDC_2019-nCoV_N2-R | GCGCGACATTCCGAAGAA | None | 0.5 mM | |
| CDC-N2-P | CDC_2019-nCoV_N2-P | ACAATTTGCCCCCAGCGCTTCAG | FAM, BHQ-1 | 0.2 mM |
Table 1: List of primer-probe sets used in this study to check sensitivity and accuracy.
The cycling conditions consisted of RT at 50° Celsius for 5 minutes, initial denaturation at 95° Celsius for 10 sec, and 45 cycles of denaturation at 95° Celsius for 5 seconds and annealing/extension at 60° Celsius for 30 seconds.
Detectability of positive and negative case
The positive cased were judged by following rules: If the detection signal arise at Ct (cycle threshold) values ≥ 20 but ≤ 35 were defined as positive cases while at Ct values >35 were defined as weak positive cases. Otherwise, all were defined as negative cases.
Results
All samples were tested simultaneously for JPN-N2, JPN-N1, CDC-N2, and CDC-N1 primer/probe set (Note: As of May 2020, the Japanese testing protocol for COVID-19 is only JPN-N2 as announced by the Japan’s NIID) (Table 2). Outlines that 85 samples (82 saliva samples and 3 nasopharyngeal swab samples) of a total 102 samples that were tested positive for JPN-N2 primer/probe set while 17 samples (15 saliva and 2 nasopharyngeal swab samples) were tested negative.
| JPN-N2 positive |
JPN-N2 negative |
Total | |
|---|---|---|---|
| Saliva | 82 | 15 | 97 |
| Nasopharyngeal swab | 3 | 2 | 5 |
| Total | 85 | 17 | 102 |
Table 2: Distribution of positive SARS-CoV-2 samples for JPN-N2.
However, Table 3 depicts that among the 17 samples which were tested negative for JPN-N2, 12 samples were tested positive for CDC-N2 primer/probe set while negative for CDC-N1, 4 samples tested positive for both CDC-N1 and CDC-N2. Furthermore, one sample was tested positive for CDC-N1 while negative for CDC-N2 (Figures 3a-3h).
| CDC-N1 positive |
CDC-N1 negative |
CDC-N1 positive |
Total | |
|---|---|---|---|---|
| CDC-N2 negative |
CDC-N2 positive |
CDC-N2 positive |
||
| Saliva | 1 | 10 | 4 | 15 |
| Nasopharyngeal swab | 0 | 2 | 0 | 2 |
| Total | 1 | 12 | 4 | 17 |
Table 3: Breakdown of JPN-N2 negative samples having positive response from CDC-N1 and/or CDC-N2.
Figure 3: The detectability of COVID-19 positive or negative cases differs depending upon the primer-probe sets used. a) A RT-PCR results for JPN-N2 and JPN-N1 primer-probe sets: JPN-N2 showed relatively higher signal intensity compared to JPN-N1. b) Testing Protocol by Genesis Healthcare incorporating various primers including Japan’s NIID testing protocol (Primers: JPN-N2, CDC-N1 and CDC-N2). Test results irregularities: c) A Japanese case that was detected negative for JPN-N2 and CDC-N1 however was detected positive for CDC-N2. d) A Japanese case that was negative for JPN-N2 however was detected positive using CDC-N1 and CDC-N2 primer-probe sets. e) A Japanese case that was detected negative for JPN-N2 and CDC-N2 while showed positive for both CDC-N1 primer-probe sets. f) A Japanese case that was positive for JPN-N2 however was detected negative by using CDC-N1 and CDC-N2 primer-probe sets. g) A Japanese case that was detected positive for CDC-N2 and week positive (low signal intensity) for JPN-N2 and negative for CDC-N1 primer-probe sets. h) A Japanese case that was detected positive for both JPN-N2 and CDC-N1 but negative for CDC-N2 primer-probe sets.
These 17 “test result irregularities”, which accounts for 16.7% of total 102 positive sample pool would be declared negative if a laboratory only tested JPN-N2. This high rate of irregular occurrence stemming from 1 primer/probe can be considered as one of the major causes leading to false negatives that are often reported in association with COVID-19 testing accuracy. Furthermore, from the observation above, a combination of multiple primers, in this case, a combination of JPN-N2 and CDC-N2 primer shows the highest rate of accuracy and sensitivity compared to JPN-N2 primer alone, followed by a triple combination of JPN-N-2, CDC-N2 and CDC-N1 for the Japanese population (Figures 3b-3d). These combinations, however, may change with various COVID-19 strains that could exist in other populations and further investigation is necessary to identify the optimum combination of primers for the Japanese population as cross-border travel brings different COVID-19 strains that react to different primers.
Sample observation of RT-PCR data of irregular samples
Below are several examples of unique and irregular samples and its RT-PCR data that could serve as future hypothesis for identifying and detecting the causes of false negative RT-PCR results.In the first case, samples reacted only to the CDC-N2 primer but not to the JPN primers. One of the most problematic cases observed are the samples whereby JPN-N2 primer resulted in negative albeit responding positive to CDC-N2 or CDC-N1 primers. Since Japanese testing protocol for COVID-19 is JPN-N2 alone, registered Japanese clinical laboratories are expected to call based on N2 primer result. However, in instances such as these, the importance to provide additional information regarding the test result for CDC is recognized and attending physicians are strongly urged to retest the patient again via RT-PCR or utilize other testing methods such as CT Scan and antigen. This would diminish the risk of misinformation and potentially spread of the infection to others for patients, whether symptomatic or asymptomatic.
Among the 4 primer-probe sets, only CDC-N2 showed positive signal at late cycle (40 x) for one case. This patient (male patient A) became symptomatic 14 days before testing and upon taking PCR test on day 5, tested positive. After 1 week of quarantine and the public health office has lifted his self-quarantine, he voluntarily retested to reconfirm his status. His results showed that his test result would be negative under Japan NIID’s JPN-N2 standalone testing protocol, but the results clearly identified that he is still positive on CDC-N2 and would be declared positive if this person were to be tested at a laboratory in the US, where CDC-N2 is the standard protocol.
The second case also reflects how JPN-N2 was negative while CDC-N1 and CDC-N2 both showed positive responses at later cycles than JPN-N2 positive control (36.54 cycles and 37.44 respectively).
Primers vs. viral genetic variations
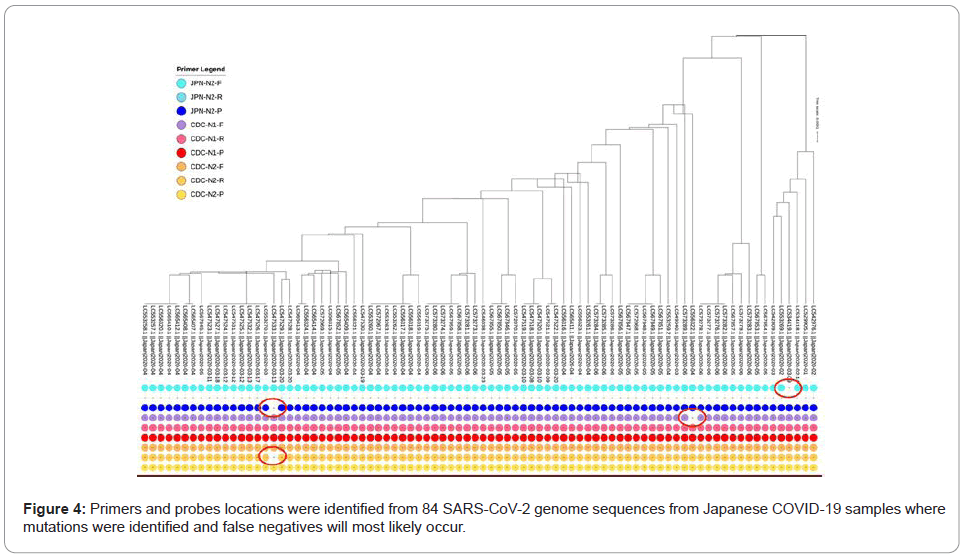
In an effort to try to explore the primer(s) effectiveness in current virus variations, an exploratory data analysis has been conducted over 84 virus strains from Japan, retrieved from NCBI GenBank genetic sequence database as of August 28, 2020.
From extracted Japan’s samples, a phylogeny tree was generated using tool from NCBI Virus [7]. The resulting tree shows different clades connected in hierarchies, suggesting that there are different virus mutations in the geographical region. Next, a sequence alignment using primers nucleotide from different countries against all coronavirus 2 strains from Japan was performed, using Blastn. The alignment results are then filtered to contain only perfect consecutive alignment of all nucleotides, meaning identities must be exactly equal to the length of primer oligonucleotide sequence with no gap. With such a rigid filter, the number of alignment matches were visualized using iTOL [8] and many virus strains those do not match with some primers are found as shown in Figure 4.
Discussion
There are numerous factors that could affect RT-PCR result for the detection of SARS-CoV-2 ranging from sample collection method, collection timing, virus inactivation technique, and many other RTPCR procedures. There are testing guidelines for COVID-19 test in many countries including NIID in Japan, CDC in USA, China, Germany and others. However, it is possible that results discrepancy occurs due to primers efficiency, sufficient viral RNA present in the samples, and due to genetic variation at the primer-probe binding sites ever after following the testing protocols and guidelines strictly. Given that Genesis Healthcare followed all protocols and guideline very strictly for sampling process, sample handling, sample transportation, preservation, RNA extraction and RT-PCR process. In order to avoid experimental error and data quality, multiple checkpoints including positive control was used. However, irregularities were observed most likely due to the variation in primer efficiency and sensitivity (Figures 3 a-3h) as well as might be due to occurring variation at the primer-probe binding sites.
It’s hypothesized that various virus strains align differently to different primers used globally due to its unique sequence. Specifically, at the time of analysis, there are 15,745 SARS-CoV-2 sequences submitted to NCBI GenBank [9] where each sequence contains different combination of variants/mutations. Therefore, to further validate our assumption, the virus mutation data and primers oligonucleotide sequences were investigated (Figure 3).
The finding also coincides with the study [10] reporting that of 33 oligonucleotides developed by different centers and shared by WHO, 79% (26) has at least one genome mutation at the primer binding sites from GISAID database containing 1,825 SARS-CoV-2 genomes.
While one dominant strain will test positive for a specific primerprobe set, there might be irregularities that will not display positive for other primer-probe sets, most likely resulting in false negative calls. In this respect, laboratories should examine their primers and probes against all known viral genome sequences to ensure that the selected primers will not result in inaccurate calling. In addition, different combination of primer-probe sets should be used, at least three or four primer-probe sets are recommended by Genesis Healthcare to reduce or overcome false negative results constraint by RT-PCR detection method.
Conclusion
The conflicting positive/negative results from the same samples that occur among different primers (JPN-N2, JPN-N1, CDC-N2, CDC-N1) outline 1) the possibility of high occurrences of false negative testing results by laboratories where JPN-N2 is the only primer used for SARS-CoV- 2, and 2) the probability that false negative/positive will likely occur when the primer binding site lies in the virus mutated location for JPN-N2 and CDC-N2.
This initial observation also suggests testing CDC-N2 in addition to JPN-2 primer, especially for symptomatic patients, with the objective to reduce false negatives and increase accuracy. Lastly, the inclusivity of CDC-N1 will result further in reducing false negatives but using CDC-N1 alone may also result in false positive calling and therefore should not be used without either JPN-N2 or CDC-N2.
Further investigation by sequencing these irregular samples will identify which primers react to different strains of the virus existing among different populations. Secondly, these irregularities raise the need for the international testing community to explore a standardized universal primer-probe sets combination for more accurate RT-PCR-based COVID-19 testing, which shall help to detect different virus strain’s reaction to selected primers by each country’s testing protocol. This is a critical consideration to ensure accurate diagnostic testing results entailing public health and safety. While more countries are re-opening their borders and resuming international travel to sustain the economic growth, this may also lead to the introduction of new strains to different populations furthering the demand for SUP.
Acknowledgement
We acknowledge the physicians from the originating medical facilities responsible for obtaining the specimen from patients.
Conflict of Interests
The authors declare that there is no conflict of interest.
Ethics Statement
Biological samples were collected from volunteers, self-medication testing participants and patients under physician care administering clinical testing for SARS-CoV-2 (COVID-19). Samples were anonymized and de-identified, so laboratory technicians and researchers were blind to the identity of the patients. Only secondary non-identifying data including age, biological sex and symptoms were provided. All experimental protocols were approved by Genesis Healthcare’s Ethics Committee.
Data Availability
The data used and produced in this research are available at:
https://github.com/genesis-healthcare/covid-19_pcr_irregularities
Funding Sources
This work is supported by Genesis Healthcare Corporation. The sponsor has no influence in the study other than providing resources and facility in conducting research and publication.
References
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, et al. (2020) Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25:2000045.
- Wu F, Zhao S, Yu B, Chen YM, Wang W, et al. (2020) A new coronavirus associated with human respiratory disease in China. Nature 579:265-269.
- Cheng HY, Jian SW, Liu DP, Ng TC, Huang WT, et al. (2020) Contact tracing assessment of COVID-19 transmission dynamics in Taiwan and risk at different exposure periods before and after symptom onset. JAMA 180:1156-63.
- Mina MJ, Parker R, Larremore DB (2020) Rethinking COVID-19 test sensitivity-A strategy for containment. N Engl J Med 383:120.
- Penarrubia L, Ruiz M, Porco R, Rao SN, Juanola-Falgarona M, et al. (2020) Multiple assays in a real-time RT-PCR SARS-CoV-2 panel can mitigate the risk of loss of sensitivity by new genomic variants during the COVID-19 outbreak. Int J Infect Dis 97:225–229.
- Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, et al. (2018) Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 34(23):4121-4123.
- Hatcher EL, Zhdanov SA, Bao Y, Blinkova O, Nawrocki EP, et al. (2017) Virus variation resource–improved response to emergent viral outbreaks. Nucleic Acids Research 45: 482-490.
- Letunic I, Bork P (2019) Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res 47: 256-259.
- Sayers EW, Cavanaugh M, Clark K, Pruitt KD, Ostell J, et al. (2020) GenBank. Nucleic Acids Res 48:84-86.
- Osorio NS, Correia-Neves M (2020) Implication of SARS-CoV-2 evolution in the sensitivity of RT-qPCR diagnostic assays. Lancet Infect Dis 21:166-167.
Citation: Tsutae W, Chaochaisit W, Aoshima H, Ida C, Miyakawa S, et al. (2021) Detecting and Isolating False Negatives of SARS-Cov-2 Primers and Probe Sets among the Japanese Population: A Laboratory Testing Methodology and Study. J Infect Dis Ther S1: 004. DOI: 10.4172/2165-7386.s1.10004
Copyright: © 2021 Tsutae W, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 2410
- [From(publication date): 0-2021 - Apr 04, 2025]
- Breakdown by view type
- HTML page views: 1661
- PDF downloads: 749