Research Article Open Access
Degradation and Colonization of Cellulose by Diazotrophic Strains of Paenibacillus polymyxa Isolated from Soil
| Ewa Beata Górska1, Urszula Jankiewicz2, Jakub Dobrzy�?�?ski1, Stefan Russel3, Stefan Pietkiewicz4, Hazem Kalaji4, Dariusz Gozdowski5 and Pawe�?�? Kowalczyk6* | |
| 1Department of Microbial Biology Faculty of Agriculture and Biology, Warsaw University of Life Sciences-SGGW, Nowoursynowska 166, 02-787 Warsaw, Poland | |
| 2Department of Biochemistry, Faculty of Agriculture and Biology, Warsaw University of Life Sciences-SGGW, Nowoursynowska 166, 02-787 Warsaw, Poland | |
| 3Institute Technology and Life Sciences in Falenty, Hrabska 3, 05-090 Raszyn, Poland | |
| 4Department of Plant Physiology, Warsaw University of Life Sciences-SGGW, Nowoursynowska 166, 02-787 Warsaw, Poland | |
| 5Department of Experimental Statistics and Bioinformatics, Faculty of Agriculture and Biology, Warsaw University of Life Sciences-SGGW, Nowoursynowska 166, 02-787 Warsaw, Poland | |
| 6Bionicum LTD, Che�?�?mska 21, 00-724, Warsaw, Poland | |
| Corresponding Author : | Pawe�?�? Kowalczyk Bionicum LTD, Che�?�?mska 21 00-724, Warsaw, Poland Tel: +48 22 840 66 99 E-mail: pawel.kowalczyk@bionicum.com.pl |
| Received November 05, 2014; Accepted January 28, 2015; Published January 30, 2015 | |
| Citation: Górska EB, Jankiewicz U, Dobrzy�?�?ski J, Russel S, Pietkiewicz S, et al. (2015) Degradation and Colonization of Cellulose by Diazotrophic Strains of Paenibacillus polymyxa Isolated from Soil. J Bioremed Biodeg 6:271. doi:10.4172/2155-6199.1000271 | |
| Copyright: © 2015 Górska EB, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
The bioconversion of cellulose to soluble sugars by diazotrophic bacteria is a very important for the environment, such as for the global stabilization and a sustainable human society. Two nitrogen-fixing microorganisms hydrolyzing cellulose were isolated from agricultural soil and identified as Paenibacillus polymyxa [the laboratory names EG2 and EG14] based on 16Sr RNA sequence. The genome of these bacteria was found to carry nif genes coding the individual components of the nitrogenase complex. Their nitrogen fixing ability was confirmed by studying nitrogenase activity in cultures of the studied bacteria in N-free medium supplemented with carboxymethylcellulose (CMC). The nitrogenase activity of P. polymyxa EG 2 was 2.9 nM C2H4?ml-1?h-1 whereas P. polymyxa EG 14 0.4nM C2H4?ml-1?h-1. The isolates in medium with filter paper synthesize following cellulolytic enzymes: carboxymethylcellulase (CMCase), FPase and Avicellase. Of the cellulolytic enzymes in the culture supernatants of the bacteria the most abundant was CMCase (P. polymyxa EG 2 103.4 mU, EG 14:96.1 mU) with far lower amounts of enzymes hydrolyzing crystalline Avicel cellulose or filter paper. In spite of these observations the better isolate in terms of synthesis of cellulases is P. polymyxa EG 14. Zymograms reflecting the main cellulase activities of the studied bacteria do not significantly differ from each other and present at least three major enzymatic activities with high molecular masses: one of about 200 kDa, another of about 220 kDa and a strong band of activity with mass of about 130 KDa. Observations of the bacterial cultures in medium with filter paper revealed the colonization of the substrate by single cells or aggregates of bacterial cells surrounded by slime. Scanning and transmission microscopy of the isolates revealed the presence of spherical structures resembling cellulosomes on the surface of the bacteria being characteristic for anaerobic bacteria of the genus Clostridium.
| Keywords |
| Degradation of cellulose; Cellulosomes; Scanning microscopy; Cellulases; Paenibacillus polymyxa |
| Introduction |
| Cellulose is a ubiquitous polysaccharide that is degraded by microorganisms belonging to various systematic groups regardless of the abiotic conditions (humidity, oxygenation, pH etc.) in the environment. Strains of bacteria belonged to the genus Paenibacillus are among the isolated microorganisms capable of hydrolyzing cellulose. |
| Paenibacillus polymyxa is a spore-forming, Gram-positive, facultaively anaerobic, heterotrophic bacterium that lives in various environments such as: soil, waters, the surface and interior of roots and the above-ground of plants [1]. It utilizes various organic and mineral compounds as a source of nitrogen. Some strains synthesize nitrogenase and are able to assimilate molecular nitrogen [2,3]. P. polymyxa produces lectins, heteroauxins, antibiotics (polymyxin) and other biologically active compounds, as a result of which it is included into plant growth promoting bacteria [4,5]. P. polymyxa strains are able to hydrolyse cellulose, both its amorphous and crystalline regions, by synthesizing a cellulase complex composed of endo- 1,4β-glucanase (3.2.1.4), exocellobiohydrolase (3.2.1.91), cellobiose phosphorylase (2.4.1.20) and cellodextrins (2.4.1.49). The main role in the degradation of cellulose is played by endo-β-1,4-glucanase and exocellobiohydrolase. Endo-β-1,4-glucanase hydrolyzes amorphous regions of cellulose to cellodextrins and cellotetrose, which are then degraded by exocellobiohydrolase to cellobiose [6-9]. |
| Some microorganisms produce exogenous cellulases located in surface structures termed cellulosomes [10-13]. Cellulosomes were first observed in Clostridium thermocellum, which degrades cellulose under thermophilic conditions [10,11]. There are only a few articles on the formation of cellulosomes or minicellulosomes on the surface of bacterial cells belonging to the genera Bacillus and Paenibacillus, which are capable of degrading cellulose [9,14,15]. |
| So far there have been few studies involving microscopic analysis regarding the colonization of cellulose by P. polymyxa bacteria. The problem is important as that the mechanism of cellulose degradation in the environment considerably differs depending on the taxonomic group of microorganisms carrying it out. Fungi do not require direct contact between the hyphae and substrate to hydrolyze cellulose, whereas in the case of bacterial cells, adhesion and the colonization of cellulose are necessary for the synthesis of cellulases to be initiated, particularly in the initial stages of cellulolysis. |
| The aim of our studies was to determine the activity of the cellulolytic enzymes produced by two diazotrophic strains of the bacterium P. polymyxa isolated from soil and to visualize cellulosomes on the surface of the studied bacteria as well as the colonization of cellulose by the bacterial cells using microscopy techniques. |
| Materials and Methods |
| The studies involved two strains of facultatively anaerobic sporeforming bacteria isolated from agriculturally used land. Based on the morphology of cells in 18-20 hour cultures of the bacteria in broth and of colonies on nutrient agar incubated at 28°C as well as on biochemical traits identified using Biomerieux API 50 CHB test the isolates were preliminarily classified to the genus Paenibacillus. To identify the isolates to species the sequence of their 16S rRNA was determined. To amplify the 16S RNA gene universal starters 27 F and 1401R were used. The matrix for PCR was genomic DNA isolated using Genomic DNA Purification Kit (Fermentas) from a culture of the studied bacteria in late logarithmic phase of growth. The purified PCR product was sequenced at the Laboratory of DNA Sequencing and Oligonucleotides Synthesis of the Institute of Biochemistry and Biophysics, Polish Academy of Sciences. The obtained nucleotide sequences were compared with those in the databases GenBank, EMBL (European Molecular Biology Laboratory) and DDBJ (DNA Data Bank of Japan) using the program BLAST (Basic Local Alignment Search Tool). |
| Nitrogenase activity of the studied isolates was determined by the acetylene reduction method in medium containing K2HPO4: 1 g, MgSO4 × 7H2O: 0.5 g; CaCO3: 5 g, H2): 1000 ml. The source of both carbon and energy was 1% carboxymethylcellulose added to the medium. The bacteria were grown in small 10 ml glass bottles into which 2.9 ml medium and 0.1 ml 24-hour bacterial culture (6 × 108 cellsâ�?�?ml-1) were introduced. After 6-day incubation at temperature 28�?�?C the bottles containing the cultures were tightly closed and 10% of the gaseous phase in the bottles was replaced with acetylene. After 24-hour incubation at 28�?�?C the amount of ethylene in the gaseous phase was measured using ATI Unicam type 610 gas chromatograph equipped with katharometer and 150 × 0.4 cm column packed with Porapak N (80-100 mesh). The carrier gas was helium. The activity of the enzyme was expressed as nM C2H4â�?�?ml-1â�?�?h-1. |
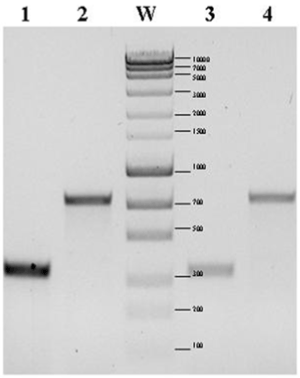
| Fixation of atmospheric nitrogen by the studied bacteria was confirmed by detection of the nitrogenase gene. To this end specific starters for amplification of a 749 bp fragment of the nitrogenase iron protein gene sequence Nitrogenase component II of P. polymyxa M1 (NC_017542) as well as to a fragment (335 bp) of nitrogenase iron protein subunit NifH P. terrae HPL-003 (NC_016641) were designed. PCR was carried out using genomic DNA as the matrix. |
| The ability of the isolated bacterial strains to synthesize exocellulases was studied in Park's mineral medium [16] supplemented with 50 mg average filtration quality filter paper by the spectrophotometric method described by Ghose [17]. The growth medium was inoculated with 0.5 ml cells of the studied isolates equivalent to a MacFarland 7 standard. Cultures of each strain were set up in triplicate and incubated 28°C. In supernatants from 14 and 28-day bacterial cultures the activity of FPase (complex of enzymes saccharifying cellulose), Avicellase and carboxymethylcellulase (CMCase) was studied. |
| The activity of FPase and Avicellase was studied in post-culture fluids by measuring the amount of reducing sugars split off by hydrolysis from 50 mg Whatman 1 filter paper or 50 mg microcrystalline Avicel cellulose respectively. Besides cellulose the reaction mix contained 0.5 ml supernatant; 0.5 ml of 0.05 M potassium phosphate buffer, pH 7.0. |
| The reference sample differed from the test samples in that it contained 3 ml DNS. Both samples were incubated at 50°C; 60 min for the FPase and 90 min for Avicellase. After incubation 3 ml DNS were added to the test samples to terminate the reaction and create a colour complex as a result of the reduction of the reagent by the reducing sugars released by hydrolysis. All the samples were then placed for 5 min in boiling water and then they were cooled to room temperature. To each sample 20 ml distilled water was added and absorbance was read against water at λ=540 nm. CMCase activity was determined similarly as for FPase and Avicellase except that Whatman 1 filter paper or microcrystalline Avicel cellulose was replaced with 2% solution of CMCose in 0.5 ml potassium phosphate buffer, pH=7.0. The reaction mixtures were incubated for 30 min at temp. 50°C. |
| One unit (U) of FPase, Avicellase and CMCase activity was defined as the amount of enzyme, which under the conditions used releases an amount of reducing sugars equal to 1 μmole of glucose in one minute. |
| To detect the enzymatic proteins in the cellulase complex the supernatants obtained after centrifuging cultures of the studied bacteria were subjected to electrophoretic separation under denaturing conditions (SDS–PAGE). |
| An 8% separating polyacrylamide gel containing 0.1% CMC and a 4% stacking gel were employed. Following electrophoresis the gels were incubated for 15 min at 50°C and then stained with a 0.1% solution of Congo red as described by Ratanakhanokchai et al. [18]. |
| Observations of the colonization of filter paper and microcrystalline Avicel cellulose in P. polymyxa cultures were carried out using laser scanning confocal microscopy at the Laboratory of Confocal Microscopy and Image Analysis of the Jagiellonian University. Cultures of the studied bacteria were incubated at 28°C in Park's medium with strips of filter paper or microcrystalline Avicel cellulose. After 6 days the cellulose together with multiplied bacteria was stained in situ with acridine orange [19] in concentration 10 μg/ml. Acridine orange is a metachromatic fluorescent stain with affinity for nucleic acids. Singlestranded nucleic acids are stained red (excitation in purple light), and double-stranded ones green (excitation in blue light) [20,21]. Acridine orange easily adsorbs on the surfaces of various materials, including cellulose. |
| The preparations were observed under a laser scanning confocal microscope using: |
| • Bio-Rad MRC 1024 system, equipped with Nikon Diaphton 300 microscope and argon laser with power 100 mW (ILT) |
| • 10 x Plan Fluor (NA 0.3) and 60 x Plan Apo (NA 1.4) lens for oil immersion |
| • VHS (51 ODCLP) and A2 (565 DRLP) filter blocks, 53 ODF 40 and 585 LP emission filters |
| • Excitation with 457 nm line. |
| Examination of bacterial cells using transmission electron microscope was as described by Vladut-Talor et al. [22] and Lamed et al. [23]. The strains were grown in Park’s medium [16] supplemented with 0.5% cellobiose at temp. 28°C. After 24 hours the culture was centrifuged and the harvested bacterial cells were treated for 2 hours with 2% glutaraldehyde solution, then washed with cacodylate buffer solution, pH 7.2 and treated for 2 hours with 2% solution of osmium tetrachloride. After this step the cells were dehydrated in a gradient of ethanol solutions and covered with synthetic resin. After drying the preparations were cut into 1-2 μm thick microsections and 80 nm thick ultrasections with LKB ultramicrotome. For light microscope observations the microsections were treated with 0.1% methylene blue in 1% borax solution. The ultrasections, on the other hand, were treated with a solution of uranyl acetate and lead citrate and then observed using JEOL JEM-1200 EX transmission electron microscope. |
| The presence of cellulosomes on the surface of Paenibacillus sp. was investigated with transmission and scanning electron microscope. For control preparations cells that had been grown in the presence of cellobiose as well as those that had not been treated with ferritin were taken n. |
| Transmission electron microscopy |
| Cellulosomes on the surface of bacterial cells were localized using a cytochemical method with cationized ferritin (CF, Sigma Chemical Co. St. Louis, USA). Following 24 hour incubation in mineral medium containing 0.5% cellobiose the bacterial cells were spun down, washed several times with physiological salt solution (0.9% NaCl) and then resuspended in a mixture of 0.7 ml 0.9% NaCl solution and 0.3 ml reagent according to Karnovsky [24]. After 20 minutes the cells were washed again in 0.9% NaCl solution. The bacterial suspension was treated with a mixture of 0.7 ml of 0.9% NaCl solution and 0.3 ml of 0.9% NaCl solution containing 0.5 mg cationized ferritin. After 1 hour the cells were washed in 0.9% NaCl solution. Cells not treated with cationized ferritin solution were the control. Further steps were as above [24,25]. |
| Scanning electron microscopy |
| A 24-hour culture in Park's mineral medium with 0.5% cellobiose was centrifuged in an Eppendorf centrifuge. The bacterial pellet was washed for 10 min. in 0.9% physiological salt solution and fixed for 20 min with Karnovsky’s reagent, pH 7.2 (in 0.1 M phosphate buffer). The cells were then washed three times 0.9% in NaCl solution and 0.7 ml of the bacterial suspension was incubated for 1 hour with a solution of cationized ferritin (0.3 ml CF contained 0.5 mg of this reagent). Following this step the cells were washed in NaCl solution and fixed once more for 20 min in Karnovsky’s reagent, then washed three times in NaCl solution and finally dehydrated in a gradient of ethanol concentrations. Finally, after being washed three times in acetone the cells were dried at critical point using liquid CO2 at 45°C and pressure 175 atm and coated with gold in Type JEE-4B evaporator manufactured in Japan. The preparations were observed with JEOL JEM-1200 EX electron scanning microscope equipped with ASID [23]. |
| Results |
| Analysis of the 16S rRNA gene sequences of both isolates with laboratory names EG2 and EG14 showed that they belong to the species P. polymyxa. The nucleotide sequences have been registered and obtained the accession numbers in DDBJ/EMBL/GenBank database AB808724 (EG2) and AB808725 (EG14), respectively. |
| The studied isolates synthesize nitrogenase and when grown in nitrogen-free medium with carboxymethylcellulose as sole carbon source cellulose were able to fix atmospheric nitrogen. Nitrogenase activity in P. polymyxa EG2 and EG14 cultures in medium supplemented with carboxymethylcellulose was 2.9 nM C2H4��?ml-1��?h-1 and 0.4 C2H4��?ml-1��?h-1, respectively. |
| The presence of genes coding the individual components of the nif cluster was confirmed by PCR using specific starters: 5’-GTTTTACGGCAAGGGCGGTATCGGCA-3’ and 5’-CCTCCAGCTCTCCATGGTGATCG-3’. The size of the obtained products of amplification confirmed the presence in the genome of both studied strains of nitrogenase iron protein subunit NifH gene (Figure 1) and nitrogenase iron protein gene (Figure 1). |
| The two studied P. polymyxa strains produced cellulases degrading both amorphous and crystalline cellulose. The isolates in medium with Whatman 1 filter paper synthesized mainly carboxymethylcellulase as well as significantly lower amounts of enzymes that hydrolyzed crystalline Avicel cellulose or filter paper. However, in spite of the similarity the better isolate in terms of synthesis of cellulases was P. polymyxa EG14 (Table 1). |
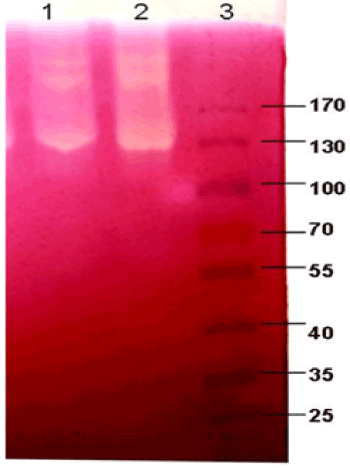
| Zymograms illustrating the main cellulase activities of P. polymyxa EG2 (well 1) and P. polymyxa EG14 (well 2) are shown in Figure 2. The zymograms for both strains did not significantly from one another. At least three main enzymatic activities with high molecular masses are visible: one of about 200 kDa, another of about 220 kDa and a strong band of activity with mass of about 130 KDa. |
| Observations of cultures of P. polymyxa strains in Park's medium containing filter paper and microcrystalline Avicel cellulose as carbon source in laser scanning confocal microscope are presented in Figures 3-5. |
| In the studied system live, active metabolically bacterial cells (bak) stained red. Acridine orange bound to cellulose (cel) was in forms showing both green and red luminescence. The proportion of these colours was different in different cellulose fibres in filter paper and in the crystalline cellulose particles. Hence the fluorescence of stained cellulose was recorded as yellow-red, yellow or yellow-green. |
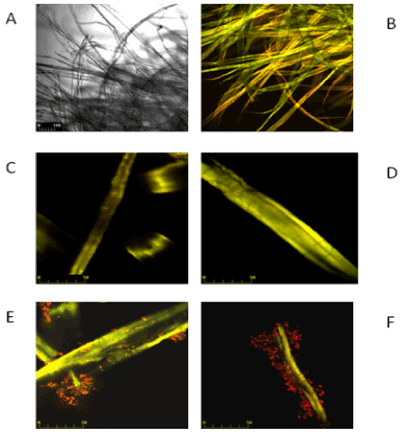
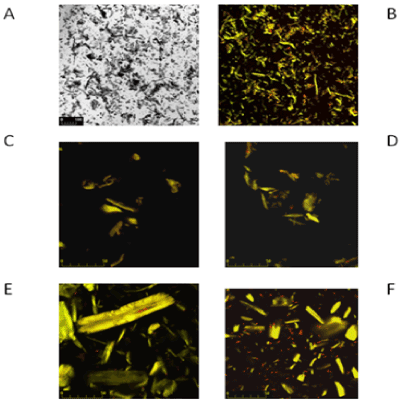
| On Figures 3 and 4 the images marked A, B, C and D are controls of filter paper and Avicel cellulose, respectively, whereas image E is cellulose with P. polymyxa AB 808 724(EG2) cells and image F presents P. polymyxa AB 808 725 (EG14) on cellulose. Non-confocal images obtained in transmitted light are designated with the symbol A, whereas B images are the same fields of vision-confocal images of acridine orange fluorescence. Images C and D are examples of the control under stronger magnification. The presented Figures 3 and 4 show the presence of bacteria on the surface of the cellulose fibres and crystals as well as in other parts of the field of vision, which indicates the colonization of the substrate-cellulose. |
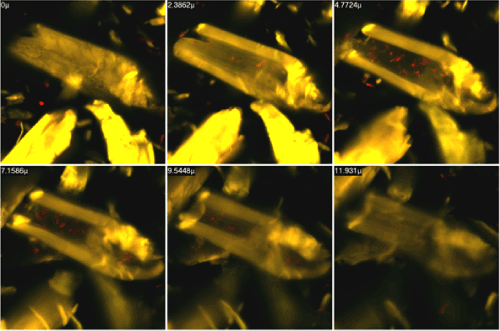
| Figure 5 presents an acridine orange stained preparation prepared from P. polymyxa AB808724 (EG2) culture in Park's medium containing microcrystalline Avicel cellulose. The individual figures were taken using laser scanning confocal microscopy of the cell surface (0 μ) and of the following layers of the preparation: 2.4 μ; 4,8 μ; 7.2 μ; 8.5 μ and 11.9 μ deep. Based on observations and computer analyses of the obtained images the penetration of bacterial cells into the Avicel cellulose crystals can be concluded. |
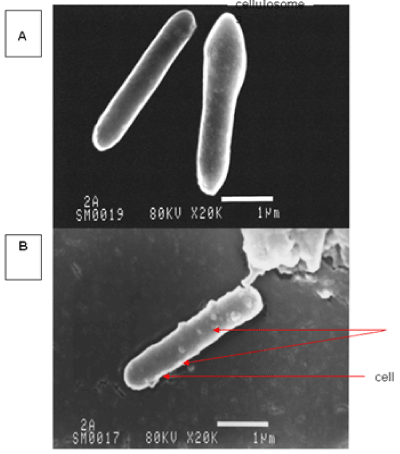
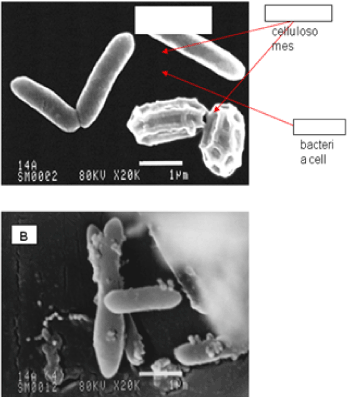
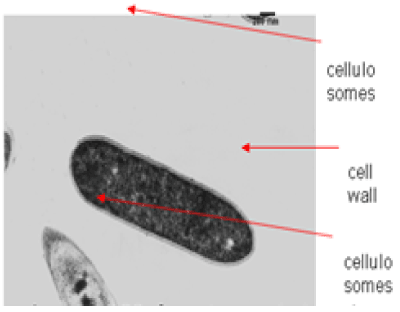
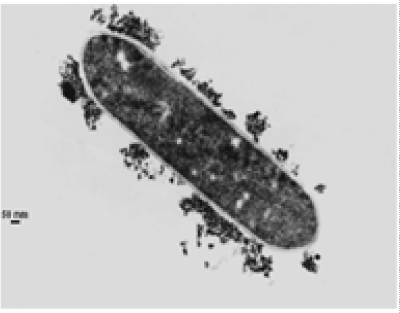
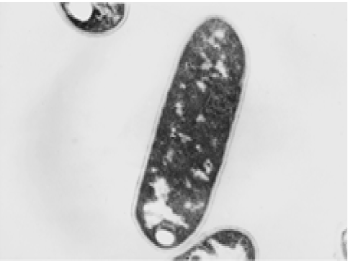
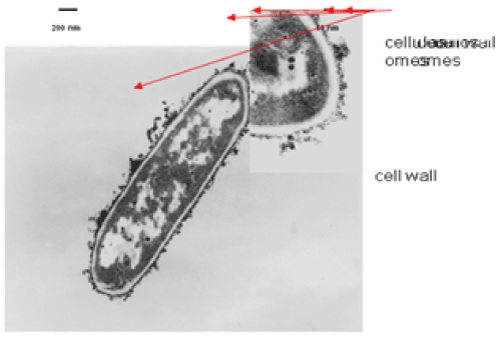
| The results of microscopic observations of cellulosomes on the surface of the cells of the studied bacterial strains are presented in Figures 6-11. The studied cells were treated with ferritin and analyzed by scanning (Figures 6b and 7b) and transmission electron microscopy (Figures 9 and 11). Around the cells not treated with ferritin and not grown with cellobiose, both in scanning and transmission electron microscope no structures, known in the literature as cellulosomes, were observed (Figures 6a, 7a, 8 and 10). |
| Figure 6b presents a scanning electron microscope image of ferritin-treated P. polymyxa AB808724 cells. On the surface of the cells „protuberances” resembling cellulosomes are visible. An analogous picture from P. polymyxa AB808725 culture is presented in Figure 7b. In preparations of P. polymyxa AB808724 (EG2) and P. polymyxa AB808725 stained with ferritin and observed in transmission microscope, around the cells irregular cellulosome structures are visible (Figures 9 and 11). These structures can be single or in aggregates, both adjacent to the bacterial cell surface or a certain distance from it. The cell surface clearly shows spherical aggregates of cellulosomes tethered to the cell surface in the form of granular growths. |
| Discussion |
| The microbiological degradation of cellulose involves the biosynthesis of many different enzymes. In order to completely break down insoluble substrates like filter paper or crystalline Avicel cellulose the cooperation of different enzymes forming a cellulase complex is necessary. These enzymes can be released by the cells and occur freely in the medium or be in free form in the medium or bound to the bacterial cell as cellulosomes. The attachment of bacterial cells to the surface of cellulose can also be mediated by slimy and yellow substances they produce [26]. Most bacteria with cellulolytic properties produce yellow substances with chemical structure similar to that of the carotenoids. These substances probably increase the affinity of cellulases for the substrate and can play an important role in the adhesion of bacterial cells to the substrate. |
| Zymograms obtained after electrophoretic separation crude enzymes (SDS PAGE) allowed detecting the activity of three cellulolytic enzymes with approximate molecular masses of 220 kDa, 200 kDa and 130 kDa. Cellulases with similar molecular masses have been described for other bacteria belonging to the genus Paenibacillus [15]. However, no activity bands with lower molecular masses, as described for P. barcinonensis [27] or P. curdlanolyticus B-6 [9], were obtained. |
| Zymograms from the studied bacteria showed only three intense bands of CMCase activity. On the other hand, the multi-enzyme complex from P. curdlanolyticus B-6 was composed of 10 CMCase activities), whereas the cellulosomes of P. sp TW1 contained 6 cellulolytic activities [28]. |
| In the sixties the first papers on the fixation of molecular nitrogen by bacteria of the genus Bacillus were published [29,30]. The studies by these authors were confirmed in the eighties by acetylene reduction methods. These studies involved the generally known bacterial species B. polymyxa (currently Paenibacillus polymyxa) and B. macerans as well as two species B. azotofixans and B. psychrosaccharolyticus not included in Bergey’s bacterial taxonomy [31]. |
| The genomes of the two P. polymyxa isolates described in this paper carry nif genes coding nitrogenase. The synthesis of this enzyme by the studied bacteria in nitrogen-free medium with carboxymethylcellulose CMC as sole carbon source confirmed the results obtained by us using the acetylene reduction method. As yet no nitrogenase activity in cultures of Paenibacillus sp. in medium with cellulose has been found but the synthesis of the enzyme in nitrogen-free medium supplemented with glucose or simple sugars has been described elsewhere [14]. |
| Microscopic observations made with laser confocal microscope of BF fibres and cellulose crystals from Paenibacillus sp. Culture in Park’s medium revealed the adherence of the bacterial cells to the cellulose. Treatment of cultures of both strains with acridine orange facilitated distinguishing live bacterial cells (red colour) from the substrate (yellow) (Figures 3-5), which allowed identifying their position with respect to the fibres of filter paper and crystals of Avicel cellulose. Microscope studies revealed colonization of the surface of cellulose by individual bacterial cells as well as the attachment of aggregates of cells bound together by slimy substances to filter paper, as shown in Figure 3. It cannot be unequivocally stated that the slime is involved in the attachment of the bacterial cells to cellulose since aggregates of cells in slime are not only on the surface of the cellulose, but also beyond it. These aggregates are not observed in culture with microcrystalline Avicel cellulose as a carbon source (Figures 4 and 5). |
| The ability of the studied isolates to hydrolyze cellulose, which is initiated by the colonization of the polysaccharide, was confirmed by enzymatic studies. The bacterial cultures grown in medium with filter paper showed cellulase activity carrying out the hydrolysis of the amorphous and crystalline regions, i.e. FPase, Avicellase and carboxymethylcellulase (CMCase) |
| The degradation of crystalline cellulose by P. polymyxa is interesting from the viewpoint of the mechanism of this process. Cellulose crystals have a spatial structure and in this connection it has been postulated that their degradation involves their penetration by bacteria. To check this assumption, Avicel cellulose crystals from a culture of P. polymyxa were analyzed layer by layer using a laser scanning confocal microscope. Microscopic images of select layers are presented in Figure 5. Analysis of these images confirms the assumption that bacterial cells penetrate the interior of the crystal. |
| In the case of some bacteria belonging to cellulolytic microorganisms the presence of multi-enzyme complexes called cellulosomes is found. A single complex may contain even a dozen or so proteins. The proteins secreted outside the bacterial cell adopt the form of spherical structures. Sometimes single cellulosomes combine to form polycellulosomes [23]. Cellulosomes usually consist of two components. One of them is a catalytic subunit with enzymatic activity, the other binds the substrate. Both components are closely linked [32]. Cellulosomes were first observed on the surface of Clostridium thermocellum cells [33]. The thermophilic Cl. thermocellum synthesizes a complex with mass over 2000 kDa. The separation of a single cellulosome revealed the presence of fourteen different proteins with molecular masses within the range from 45 kDa to 210 kDa. Such a large number of polypeptides is determined by the presence of genes coding these enzymes, e.g. in Cl. thermocellum NC1B 10682 there are15 genes coding endoglucanases, 2 genes responsible for the expression of xylanases, 2 genes coding cellobiase and one coding lichenase [34]. Furthermore, Bagnara-Tardif et al. [35] isolated from the genome of Cl. thermocellum gene celS coding a cellobiohydrolase with significant activity against carboxymethylcellulose. |
| The best characterized cellulosomes are those from the surface bacteria belonging to the genus Clostridium. The properties of these multi-enzyme complexes have been studied in C. thermocellum, C. cellulolyticus and C. cellulovorans [10,12,13]. Cellulosomes have also been discovered in non-endospore forming bacteria belonging to the genera Acetovibrio, Butyrivibrio, Bacteroides, Ruminococcus and Streptomyces [36]. |
| Cellulolytic and hemicellulolytic protein aggregates with enzymatic activities have also been described for mesophilic bacteria of the genera Bacillus and Paenibacillus. So far cellulosomes from B. circulans [37], B. megaterium [6], B. polymyxa [14] and P. curdlanolyticus [9] have been isolated and described. |
| Observations made using scanning and transmission microscopy of the cell surface of the studied cellulolytic strains of P. polymyxa treated with ferritin confirmed the presence on their outer surface of spherical structures occurring single or in complexes. Some of them were outside the cell. These structures resemble the cellulosomes and polycellulosomes described in the literature. Lamed et al. [23] and Mayer et al. [33] demonstrated that isolated and purified cellulosomes are capable of hydrolyzing cellulose in vitro to cellobiose. |
| In 1993 Kim and Kim [37] identified two cellulosomes in Bacillus circulans. Both complexes degraded xylan, filter paper, CMC, cellobiose, microcrystalline Avicel cellulose, cotton and p-nitrophenyl-β-Dcellobiose, whereas an isolated and purified enzyme with molecular mass 75 kDa successively degraded Avicel cellulose. The molecular mass of the enzyme complexes C-1 and C-2 was 669 and 443 kDa. Complex C-2 consisted of at least five enzymes degrading CMC and two xylanases, whereas C-1 had three CMCases and four xylanases. In complex C-1 the highest activity was shown by β-glucosidase, CMCase, xylanase and cellobiohydrolase, and in C-2 avicellase, β-glucosidase, CMCase and xylanase. |
| Waeonukul et al. [9] isolated a multi-enzyme complex from P. curdlanolyticus B-6 during cultivation on microcrystalline Avicel cellulose under aerobic conditions. A single cellulosome was capable of hydrolyzing both Avicel cellulose as well as insoluble xylan. Moreover, it was found that this complex also contained avicellase, CMCase, cellobiohydrolase, β-glucosidase, xylanase, β-xylosidase and α–Larabinofuranosidase. The total mass of the multicomplex was ca. 1600 kDa. The complex was composed of twelve proteins, of which ten were able to degrade carboxymethylcellulose. The isolated cellulosome efficiently degraded lignocellulose. |
| Van Dyk et al. [32] and Mayer et al. [33] demonstrated that B. licheniformis strain SVD1 synthesizes multi-enzyme complexes (MEC) with hemi-cellulolytic activity. The total molecular mass of the complex was approximately 2000 kDa. Other enzymes forming MEC hydrolyzed xylan, mannose, pectin and carboxymethylcellulose. However, no avicellase activity was found, which precludes calling this multi-enzyme complex a cellulosome. However, it is possible to use genetic modifications that would allow the use of the MEC in many areas of the biodegradation of natural wastes. |
| Conclusion |
| • The investigated spore-forming bacteria belong to the species P. polymyxa. |
| • The two strains P. polymyxa are diasotrophic bacteria because showing of nitrogen fixing ability. |
| • These isolates can to degradate the amorphous and crystalline regions in cellulose. |
| • The cells of investigated bacteria colonise of cellulose fibers during the degradation. |
| These bacteria when cultured in medium with filter paper form spherical structures resembling cellulosomes on the surface of the bacteria such as anaerobic, cellulolytic bacteria of the genus Clostridium. |
References
|
Tables and Figures at a glance
| Table 1 |
Figures at a glance
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
 |
 |
 |
 |
| Figure 5 | Figure 6 | Figure 7 | Figure 8 |
 |
 |
 |
|
| Figure 9 | Figure 10 | Figure 11 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 15439
- [From(publication date):
March-2015 - Apr 04, 2025] - Breakdown by view type
- HTML page views : 10728
- PDF downloads : 4711
