Review Article Open Access
Current Methods for Analysis of Enzymatic Peptidyl-tRNA Hydrolysis
Hana McFeeters and Robert L McFeeters*Department of Chemistry, University of Alabama in Huntsville, USA
- *Corresponding Author:
- Dr. Robert L McFeeters
Associate Professor, Department of Chemistry
University of Alabama in Huntsville
301 Sparkman Dr, Huntsville, AL 35899, USA
Tel: (256) 824-6023
Fax: (256) 824-6349
E-mail: robert.mcfeeters@uah.edu
Received date: September 15, 2014; Accepted date: October 28 2014; Published date: October 31, 2014
Citation: McFeeters H, McFeeters RL (2014) Current Methods for Analysis of Enzymatic Peptidyl-tRNA Hydrolysis. J Anal Bioanal Tech 5:215. doi: 10.4172/2155-9872.1000215
Copyright: © 2014 McFeeters H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
Understanding peptidyl-tRNA and the enzymes responsible for recycling them has come from the ability to detect and quantify enzymatic peptidyl-tRNA hydrolysis. The methods available to study removal of peptides from tRNA have evolved considerably. Radioactive [14C] amino acids were first implemented to monitor cleavage of the peptide-nucleotide ester bond of uniform peptidyl-tRNA substrates. Later, Northern blots with radiolabeled oligonucleotide probes were used to observe cleavage of specific peptidyl-tRNAs or individual tRNA from bulk peptidyl-tRNA populations. Finally, the use of fluorescently labeled amino acids was introduced, which could be coupled to anisotropy or PAGE readouts. Here we review the methods for quantification and analysis of enzymatic peptidyl-tRNA hydrolysis and summarize their inherent advantages and disadvantages.
Keywords
Peptidyl-tRNA; Peptidyl-tRNA hydrolase; Enzyme hydrolysis; Functional assay; Pth1; Pth2
Introduction
Peptidyl-tRNAs are thought to be inconspicuous molecules found in all cells. This is mainly due to cells having developed efficient means to recover tRNA from the unusable peptide bound form. However, the significance of peptidyl-tRNA and its enzymatic hydrolysis is finally being discovered, particularly in regard to impact on protein biosynthesis and cell viability.
In the course of translation, ribosomes stall on average 10% of the time [1]. If not rescued, disassembly of the translation machinery results in the release of peptidyl-tRNA. Peptidyl-tRNA is also generated from translation of minigenes or short ORFs [2-4]. Build-up of peptidyl-tRNA is lethal due to tRNA starvation [5]. The importance of preventing peptidyl-tRNA accumulation is apparent from the multiple mechanisms cells employ to recycle them [6,7]. These mechanisms depend on whether peptidyl-tRNA is part of the stalled ribosome or has been released. In stalled ribosomes, peptidyl-tRNA is recycled by ribosome associated proteins with peptidyl-tRNA hydrolase function [8,9]. If the ribosome is disassembled and peptidyl-tRNA is released, the complexity of tRNA recycling mechanisms differs greatly between prokaryotes and eukaryotes. In most bacteria there is a single, essential peptidyl-tRNA hydrolase enzyme, Pth1, responsible for removing the peptide from peptidyl-tRNA [10]. In certain bacterial species, a Pth2 homolog has been identified [11] but has not been structurally or functionally characterized. In eukaryotes, there is an emerging network of enzymes that recycle peptidyl-tRNAs, including mitochondrial associated Pths and Pth domain containing proteins [12-14].
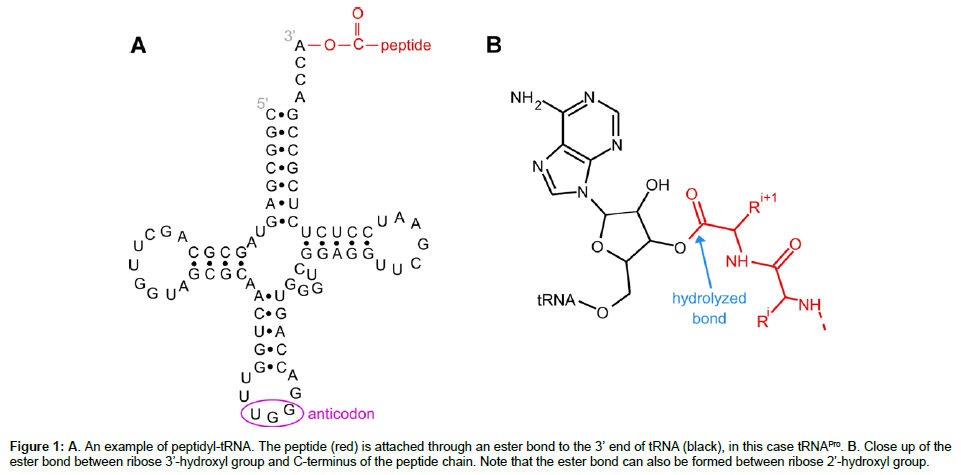
From a biochemical point of view, peptidyl-tRNA is somewhat of an enigma having dual character – part peptide, part nucleic acid, Figure 1. This dual character leads to tremendous heterogeneity. Peptidyl-tRNA isolated from any given cell is a mixture of more than 40 tRNA molecules and considerably larger number of peptides [15]. Even with all of the possible differences, the character of the tRNA component is fairly uniform – a typical “clover leaf” structure with large negative charge. Each has an attached peptide of variable length and composition. The nature of the peptide component is much more heterogeneous and largely determined by size and composition. The presence of a shorter peptide does not change the properties of the molecule much from the properties of tRNA itself. A larger peptide chain can, however, significantly alter the molecular properties and have implications for the choice of methods to analyze peptidyl-tRNA enzymatic hydrolysis.
Figure 1: A. An example of peptidyl-tRNA. The peptide (red) is attached through an ester bond to the 3’ end of tRNA (black), in this case tRNAPro. B. Close up of the ester bond between ribose 3’-hydroxyl group and C-terminus of the peptide chain. Note that the ester bond can also be formed between ribose 2’-hydroxyl group.
Herein we review the methods that have been developed to monitor the cleavage of the peptide-tRNA ester bond of peptidyltRNA. The methods are separated into three categories based on the underlying principle used to determine the degree of hydrolysis using: (1) radioactive labeled amino acids, (2) electrophoretic separation of tRNA, and (3) fluorophore conjugated amino acids. The methods differ in their requirements for peptidyl-tRNA uniformity. While most employ a specific peptidyl-tRNA, it should be noted that production of uniform specific peptidyl-tRNA molecules is rather challenging. Therefore, specific peptidyl-tRNA tends to be replaced by a modified aminoacyl-tRNA for in vitro experiments. Moreover, there are instances in which observation of a heterogeneous sample of peptidyltRNA is advantageous. The selection of an appropriate method for peptidyl-tRNA hydrolysis should take into account several factors: hydrolysis product of interest (peptide or tRNA), amount of peptidyltRNA necessary to accomplish the study, and feasibility of the method given constraints on time, equipment, and resources.
Radioactive Labeled Amino Acids
The first reported methods for study of peptidyl-tRNA hydrolysis utilized [14C] labeling. Initially a [14C] labeled amino acid was attached to the tRNA using the appropriate tRNA synthetase. Peptidyl-tRNA was then formed by condensation of an additional peptide with the [14C]-aminoacyl-tRNA [16,17]. The preparation of such peptidyltRNA was formidable but yielded a natural and very relevant substrate for enzyme studies. To measure activity, tRNA was precipitated with TCA after incubation with Pth1. Soluble hydrolysis products, the [14C] peptides, were separated with paper chromatography, visualized using ninhydrin staining, and quantified by scintillation counting of the excised peptide band. Early initial insight into the enzymatic activity of bacterial Pth1 was gained using this methodology [16,17]. Several subsequent modifications have been reported. The amount of hydrolyzed peptide was determined using scintillation of the supernatant obtained by centrifugation of the TCA quenched reaction mixture omitting the paper chromatography separation step [18] and N-acetyl-[14C]-aminoacyl-tRNA was used instead of peptidyltRNA [19]. While Pth1 is not able to hydrolyze aminoacyl-tRNA, it does catalyze the cleavage of N-acetyl-aminoacyl-tRNA. Introducing N-acetylated-aminoacyl-tRNA thus simplified the preparation of the substrate and, like the omission of the paper chromatography step, expedited the assay.
Radioactively labeled peptidyl-tRNA has also been generated using an in vitro translation system with [35S]-Methionine incorporated into TnaC peptide [20]. This peptidyl-tRNA has not been used as a substrate for Pth activity assays but, made in sufficient quantity, would be an excellent specific substrate. In addition, this system could be readily modified for production of other specific peptidyl-tRNA as in the case of fluorescently labeled peptidyl-tRNA below. Upon enzymatic hydrolysis, the remaining nonhydrolyzed substrate as well as the hydrolyzed peptide could be precipitated, separated using PAGE, and visualized by autoradiography. Quantification of hydrolysis would then be determined based on band intensity.
The methods employing radioactively labeled amino acids are well suited for determination of Pth enzyme kinetic parameters. They are extremely sensitive, can be used for a wide range of signal intensities (i.e. substrate concentrations), and quantification can be readily performed. On the other hand, they require the use of a specific peptidyl-tRNA, production of which is a multistep process that may be unnecessarily demanding for certain applications. The use of radioactivity may also be undesirable or problematic. It has also been shown that the rate of Pth1 hydrolysis depends on peptide composition [18] and length [21] which is not accounted for by using N-acetyl-aminoacyl-tRNA or a single peptidyl-tRNA species.
Electrophoretic Separation of tRNA
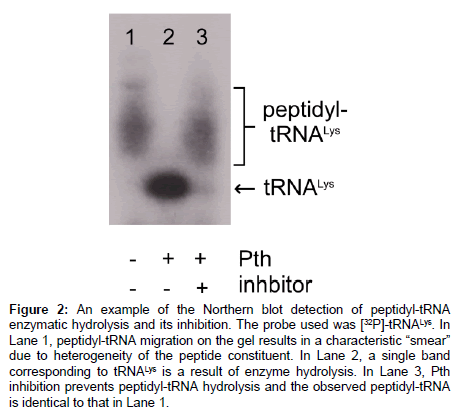
Separation of tRNA using low pH urea polyacrylamide gels was introduced to study the levels of aminoacylation in E. coli [22]. Total RNA isolated from the cell was separated on an acid-urea polyacrylamide gel and tRNAs (free, aminoacyl, and peptidyl) detected using a specific nucleotide probe labeled with [32P]-ATP (i.e. a Northern blot), Figure 2. This technique was further simplified by using minigels instead of large gels traditionally used for nucleic acid separation [23]. The analysis could be further expedited by employing nonspecific nucleotide staining [24,25]. Compared to those employing radioactively labeled substrate, this method makes observation of both the hydrolyzed product (tRNA) and the nonhydrolyzed peptidyl-tRNA possible on one gel. However, it is limited to the use of peptidyl-tRNA with very short peptides because, in general, larger peptides tend to have more positive charge at lower pH. This prevents peptidyl-tRNA from entering the gel.
Figure 2: An example of the Northern blot detection of peptidyl-tRNA enzymatic hydrolysis and its inhibition. The probe used was [32P]-tRNALys. In Lane 1, peptidyl-tRNA migration on the gel results in a characteristic “smear” due to heterogeneity of the peptide constituent. In Lane 2, a single band corresponding to tRNALys is a result of enzyme hydrolysis. In Lane 3, Pth inhibition prevents peptidyl-tRNA hydrolysis and the observed peptidyl-tRNA is identical to that in Lane 1.
Methods using electrophoretic separation of tRNA are well suited for determination of Pth enzymatic activity. They allow the use of heterogeneous peptidyl-tRNA directly isolated from cells and hydrolysis for the entire population. Specific peptidyl-tRNAs can also be used. The fact that these methods are relatively nondemanding in terms of substrates, yet work with natural substrates, makes them very attractive for many applications. The ability to perform enzymatic hydrolysis with heterogeneous, bulk peptidyl-tRNA isolated from bacterial cells, separate the reaction products on a minigel, and stain for detection makes the analysis of peptidyl-tRNA enzymatic hydrolysis achievable in any biochemistry laboratory.
Fluorophore Conjugated Amino Acids
The most recent development in the analysis of enzymatic peptidyltRNA hydrolysis has been the introduction of fluorophores conjugated to amino acids. The concept is the same as for [14C] studies but is less hazardous and has extended utility from the ability to detect changes in fluorescence anisotropy. Initial reports of fluorescent peptidyl-tRNA hydrolysis used BODIPY® FL succinimidyl ester for the labeling of lysyl-tRNALys used in downstream applications [26]. Similarly, Oregon Green was used for labeling of methionyl-tRNAMetm [27]. Later, introduction of fluorophore labeled methionine to initiator tRNA was utilized to introduce a fluorescent label to any peptide produced using an in vitro translation system [28].
Several possibilities exist to determine the amount of hydrolyzed peptidyl-tRNA using fluorescent amino acids. Peptides could be separated from peptidyl-tRNA via HPLC with a fluorescence detector measuring output [26]. Also, changes in fluorescence anisotropy could be used as an indicator of hydrolysis [26], a method adaptable to higher throughput screening. In addition, separation of hydrolysis products can also be achieved using three-layer low pH Tris-Acetate SDS-PAGE [29]. The advantage of these gels is that they enable use of peptidyl-tRNA with longer peptide constituents. Like the previous methods employing electrophoretic separation, this technique allows observation of both hydrolysis product (in this case the peptide) as well as the substrate (nonhydrolyzed peptidyl-tRNA) (Table 1).
| Detected Molecule | [Ref.] | Detection Method | Separation Technique | Sensitivity/ Quantification | Advantages and Disadvantages |
|---|---|---|---|---|---|
| Methods employing radioactively labeled amino acids | |||||
| [14C]-peptide/ N-acetyl-[14C]-leucine |
[16,17] | Ninhydrin staining/ Scintillation | Paper electrophoresis | High/Yes | (+) low amount of substrate (-) requires synthetic steps to prepare substrate or uses acetylated aminoacyl-tRNA |
| [14C]-peptide/ [14C]-diacetyl-lysine |
[18,19] | Scintillation | Centrifugation | High/Yes | (+) low amount of substrate (-) requires synthetic steps to prepare substrate or uses acetylated aminoacyl-tRNA |
| Methods employing electrophoretic separation of tRNA | |||||
| tRNA | [22,23] | [32P]-ATP (Northern blot) |
PAGE | High/Yes | (+) allows use of specific and heterogeneous substrates (-) time consuming (-) requires short peptide length |
| tRNA | [24,25] | Methylene blue Silver staining |
PAGE | Moderate/Yes High/Yes |
(+) allows use of specific and heterogeneous substrates (+) fast (-) sensitivity (-) requires short peptide length |
| Methods employing fluorescently labeled amino acids | |||||
| Fluorophore (conjugated to peptide) |
[29] | Fluorescence | SDS-PAGE | High/Yes | (+) sensitivity (-) involves extra step of fluorophore conjugation |
| Fluorophore (conjugated to amino acid) |
[26] | Fluorescence anisotropy Fluorescence |
(HPLC)a | High/Yes | (+) sensitivity (+) high throughput (-) involves extra step of fluorophore conjugation (-) uses aminoacyl-tRNA substrate |
aHPLC with a fluorescent readout was used to validate the enzymatic hydrolysis of peptidyl-tRNA while fluorescence anisotropy was used for determining the activity of Pth1.
Table 1: Summary of methods for analysis of peptidyl-tRNA hydrolysis presented here.
The development of fluorescent labeling techniques resulted in the first high throughput, anisotropy based assay for screening of potential Pth1 inhibitors [26]. Fluorescent approaches rely on fluorescently labeled specific peptidyl-tRNAs or aminoacyl-tRNA. Disadvantages to this method are high cost and batch to batch variability. On the other hand, thanks to the high sensitivity of fluorescent probes, relatively small amounts of substrate are required for detection, partially offsetting the cost. Fluorescent probes enable straightforward quantification of peptidyl-tRNA hydrolysis and Pth activity or inhibition provided the equipment (fluorescence reader, spectrofluorimeter capable of anisotropy measurements) is available. It should be taken into account that fluorescence background from natural products and synthetic small molecules can be problematic for Pth inhibitor screening.
Discussion
The development of Pth functional assays has allowed implementation of several significant applications. Having a functional assay can be used to determine whether a potential Pth gene or putative Pth-like domain can hydrolyze or even bind peptidyl-tRNA (i.e. fluorescence anisotropy). This ability has led to the definitive determination that plant CRS2 (a putative Pth) can bind, but not hydrolyze peptidyl-tRNA (McFeeters, unpublished), in agreement with in vivo complementation studies [30].
The first functional radioactive assay allowed for the determination of Pth kinetic parameters [16,21] providing insight into mechanism and comparison between different Pth enzymes or mutants [19]. A more readily producible probe for Northern blot readout enabled initial screening of Pth1 inhibitors [31,32]. Further methodological improvements, in particular the ability to use fluorescence readouts [26] has led to higher throughput screening and more potential to advance Pth1 as an antibacterial target.
In conclusion, multiple methods exist for the measurement of peptidyl-tRNA cleavage and Pth enzyme activity. Each has distinct advantages, which should be taken into account for new applications. Improvements in generation of specific peptidyl-tRNA, including synthetic approaches [33] and in vitro translation [20,28], continue to advance. With ever improving bioanalytical techniques, even simpler, more robust, and relatively quicker methods with higher throughput are on the horizon.
References
- Heurgué-Hamard V, Karimi R, Mora L, MacDougall J, Leboeuf C, et al. (1998) Ribosome release factor RF4 and termination factor RF3 are involved in dissociation of peptidyl-tRNA from the ribosome. EMBO J 17: 808-816.
- Cruz-Vera LR, Hernandez-Ramon E, Perez-Zamorano B, Guarneros G (2003) The rate of peptidyl-tRNA dissociation from the ribosome during minigene expression depends on the nature of the last decoding interaction. J Biol Chem 278: 26065-26070.
- Hernández-Sánchez J, Valadez JG, Herrera JV, Ontiveros C, Guarneros G (1998) lambda bar minigene-mediated inhibition of protein synthesis involves accumulation of peptidyl-tRNA and starvation for tRNA. EMBO J 17: 3758-3765.
- Tenson T, Herrera JV, Kloss P, Guarneros G, Mankin AS (1999) Inhibition of translation and cell growth by minigene expression. J Bacteriol 181: 1617-1622.
- Heurgué-Hamard V, Mora L, Guarneros G, Buckingham RH (1996) The growth defect in Escherichia coli deficient in peptidyl-tRNA hydrolase is due to starvation for Lys-tRNA(Lys). EMBO J 15: 2826-2833.
- Giudice E, Gillet R (2013) The task force that rescues stalled ribosomes in bacteria. Trends Biochem Sci 38: 403-411.
- Sharma S, Kaushik S, Sinha M, Kushwaha GS, Singh A, et al. (2014) Structural and functional insights into peptidyl-tRNA hydrolase. Biochim Biophys Acta 1844: 1279-1288.
- Handa Y, Inaho N, Nameki N (2011) YaeJ is a novel ribosome-associated protein in Escherichia coli that can hydrolyze peptidyl-tRNA on stalled ribosomes. Nucleic Acids Res 39: 1739-1748.
- Gagnon MG, Seetharaman SV, Bulkley D, Steitz TA (2012) Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome. Science 335: 1370-1372.
- Das G, Varshney U (2006) Peptidyl-tRNA hydrolase and its critical role in protein biosynthesis. Microbiology 152: 2191-2195.
- Powers R, Mirkovic N, Goldsmith-Fischman S, Acton TB, Chiang Y, et al. (2005) Solution structure of Archaeglobus fulgidis peptidyl-tRNA hydrolase (Pth2) provides evidence for an extensive conserved family of Pth2 enzymes in archea, bacteria, and eukaryotes. Protein Sci 14: 2849-2861.
- Dujeancourt L, Richter R, Chrzanowska-Lightowlers ZM, Bonnefoy N, Herbert CJ (2013) Interactions between peptidyl tRNA hydrolase homologs and the ribosomal release factor Mrf1 in S. pombe mitochondria. Mitochondrion 13: 871-880.
- Richter R, Rorbach J, Pajak A, Smith PM, Wessels HJ, et al. (2010) A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome. EMBO J 29: 1116-1125.
- De Pereda JM, Waas WF, Jan Y, Ruoslahti E, Schimmel P, et al. (2004) Crystal structure of a human peptidyl-tRNA hydrolase reveals a new fold and suggests basis for a bifunctional activity. J Biol Chem 279: 8111-8115.
- Dong H, Nilsson L, Kurland CG (1996) Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol 260: 649-663.
- De Groot N, Groner Y, Lapidot Y (1969) Peptidyl-tRNA. VII. Substrate specificity of peptidyl-tRNA hydrolase. Biochim Biophys Acta 186: 286-296.
- Cuzin F, Kretchmer N, Greenberg RE, Hurwitz R, Chapeville F (1967) Enzymatic hydrolysis of N-substituted aminoacyl-tRNA. Proc Natl Acad Sci U S A 58: 2079-2086.
- Shiloach J, Lapidot Y, de Groot N (1975) The specificity of peptidyl-tRNA hydrolase from E. coli. FEBS Lett 57: 130-133.
- Schmitt E, Mechulam Y, Fromant M, Plateau P, Blanquet S (1997) Crystal structure at 1.2 A resolution and active site mapping of Escherichia coli peptidyl-tRNA hydrolase. EMBO J 16: 4760-4769.
- Cruz-Vera LR, Rajagopal S, Squires C, Yanofsky C (2005) Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol Cell 19: 333-343.
- Shiloach J, Bauer S, de Groot N, Lapidot Y (1975) The influence of the peptide chain length on the activity of peptidyl-tRNA hydrolase from E. coli. Nucleic Acids Res 2: 1941-1950.
- Varshney U, Lee CP, RajBhandary UL (1991) Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J Biol Chem 266: 24712-24718.
- Janssen BD, Diner EJ, Hayes CS (2012) Analysis of aminoacyl- and peptidyl-tRNAs by gel electrophoresis. Methods Mol Biol 905: 291-309.
- Brown T, Mackey K (2001) Analysis of RNA by northern blot hybridization. Curr Protoc Hum Genet Appendix 3: Appendix 3K.
- Igloi GL (1983) A silver stain for the detection of nanogram amounts of tRNA following two-dimensional electrophoresis. Anal Biochem 134: 184-188.
- Bonin PD, Erickson LA (2002) Development of a fluorescence polarization assay for peptidyl-tRNA hydrolase. Anal Biochem 306: 8-16.
- Goodall JJ, Chen GJ, Page MG (2004) Essential role of histidine 20 in the catalytic mechanism of Escherichia coli peptidyl-tRNA hydrolase. Biochemistry 43: 4583-4591.
- Bakke CK, Jungbauer LM, Cavagnero S (2006) In vitro expression and characterization of native apomyoglobin under low molecular crowding conditions. Protein Expr Purif 45: 381-392.
- Hughes RC, McFeeters H, Coates L, McFeeters RL (2012) Recombinant production, crystallization and X-ray crystallographic structure determination of the peptidyl-tRNA hydrolase of Pseudomonas aeruginosa. Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 1472-1476.
- Ostheimer GJ, Hadjivassiliou H, Kloer DP, Barkan A, Matthews BW (2005) Structural analysis of the group II intron splicing factor CRS2 yields insights into its protein and RNA interaction surfaces. J Mol Biol 345: 51-68.
- Harris SM, McFeeters H, Ogungbe IV, Cruz-Vera LR, Setzer WN, et al. (2011) Peptidyl-tRNA hydrolase screening combined with molecular docking reveals the antibiotic potential of Syzygium johnsonii bark extract. Nat Prod Commun 6: 1421-1424.
- McFeeters H, Gilbert MJ, Thompson RM, Setzer WN, Cruz-Vera LR, et al. (2012) Inhibition of essential bacterial peptidyl-tRNA hydrolase activity by tropical plant extracts. Nat Prod Commun 7: 1107-1110.
- Graber D, Moroder H, Steger J, Trappl K, Polacek N, et al. (2010) Reliable semi-synthesis of hydrolysis-resistant 3'-peptidyl-tRNA conjugates containing genuine tRNA modifications. Nucleic Acids Res 38: 6796-6802.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 16946
- [From(publication date):
November-2014 - Apr 08, 2025] - Breakdown by view type
- HTML page views : 12322
- PDF downloads : 4624