Coronaviruses and the Associated Potential Therapeutics for the Viral Infections
Received: 21-Feb-2020 / Accepted Date: 02-Mar-2020 / Published Date: 09-Mar-2020 DOI: 10.4172/2332-0877.1000417
Abstract
Coronaviruses (CoVs), named after the crown-like spikes on their surfaces, belong to the RNA virus family Coronaviridae. Coronaviruses are enveloped RNA viruses with a linear positive-sense single–stranded genome consisting of the open reading frames (ORFs), the 5’- methylated cap, and the 3’- poly (A) tail. The open reading frames encode structural proteins and nonstructural proteins that may be drug-targetable. Antiviral medications seek to target these key proteins via different mechanisms. There have been seven coronaviruses discovered that are capable of infecting human beings, three of which including MERS-CoV, SARS-CoV and SARS-CoV-2, originated in wild animals and evolved to develop rapid human-to-human transmission, resulting in severe respiratory diseases among other pathologies, even deaths. Particularly, the outbreak of the novel Wuhan coronavirus SARS-CoV-2 has now rapidly spread across China and into many other countries. Sadly, there is no any effective vaccine or specific drug available. Many patients infected with SARS-CoV-2 have succumbed. Thus, there is a burning need for the anti-coronavirus drug development. The antiviral prodrug, Remdesivir, developed by the company Gilead, is currently under clinical investigation in China.
Keywords: Coronavirus; Structural proteins; Non-structural proteins; Infection; Drug; Vaccine
Introduction
During December 2019, patients with pneumonia of unknown symptoms first appeared in Wuhan, China [1]. And later, the novel coronaviruses (CoVs) were diagnosed and isolated and called as 2019- nCoV (or COVID-19), with a new name SARS-CoV-2 given by World Health Organization (WHO) on February 11 [2-6]. SARS-CoV-2 spread in China rapidly with person-to-person transmission. Outside of China, patients with SARS-CoV-2 have been diagnosed in 26 other countries such as Japan, South Korea, Singapore, United States, Canada and Europe. Infection with SARS-CoV-2 can result in severe diseases and even death. As of February 14, 2020, there are 66577 patients diagnosed in China, with 1524 deaths, with the mortality rate approximated at 2.29%. Meanwhile, 525 cases and 2 deaths have been reported in other 26 countries. The SARS-CoV-2 outbreak in China is currently catching global attention and is becoming a severe public health emergency.
Coronaviruses (CoVs) were first discovered in chickens in 1937 and first identified in human in 1965, and were named due to the crown- like spikes on the surface of these viruses. They belong to the RNA virus family Coronaviridae [7]. The Coronaviridae family has been divided into four genera named alpha (α) coronavirus, beta (β) coronavirus, gamma (γ) coronavirus and delta (γ) coronavirus, respectively [7,8]. There are seven coronaviruses discovered capable of infections in humans. The novel Wuhan coronavirus SARS-CoV-2 is a type of beta coronavirus.
Scientists worldwide are fighting against coronaviruses and the severe health problems associated with it. Unfortunately, there is still no effective vaccine or coronavirus-targeted therapeutic drug available, though promising progress has been made in the study of coronaviruses. Remdesivir, an anti-viral drug developed by the company Gilead, was found for its therapeutic activity against coronavirus and other RNA viruses, and is expected to be a potential effective drug. It is currently used to treat CoV patients under clinical trials in China. In this review, we will discuss the progress in CoV research and the potential development of therapeutic drugs, especially, the updated situation of SARS-CoV-2 and the possibility of the CoV-targeting drugs aimed on the CoV structural proteins.
The Classification of Coronaviruses
Coronaviruses (CoVs) only infect with human beings and other vertebrates and are associated with multiple infectious diseases, particularly, by targeting the respiratory, digestive and nervous systems. Coronaviruses (CoVs) belong to the family of Coronaviridaes and the subfamily of Orthocoronavirinaes. They are divided into four genera named alpha (α) coronavirus, beta (β) coronavirus, gamma (γ) coronavirus and delta (γ) coronavirus, respectively [9,10]. Currently, there are seven coronaviruses (CoVs) discovered to be capable of infecting human beings. Four of which only cause the common cold, including HCoV-229E (discovered in 1966), HCoV-NL63 (in 2004), HCoV-OC43 (in 1967) and HCoV-HKU1 (in 2005), with the first two being alpha (α) coronaviruses and the last two being beta (β) coronaviruses. The three other coronaviruses cause more severe respiratory disease processes. They originate in animals and eventually infect humans as they develop further. These viruses are the MERS- CoV (discovered in 2012), SARS-CoV (in 2002) and SARS-CoV-2(in 2019) [11]. All of the three belong to beta (β) coronaviruses [12,13]. As demonstrated, SARS-CoV originated from bats, and MERS-CoV was isolated from camel, but the original host of SARS-CoV-2 is still not clear.
The Genomic Structure of Coronaviruses
Coronaviruses (CoVs) are the type of enveloped RNA viruses with a linear positive-sense single– strand. These coronaviruses have large RNA genomes with the length of 27-32 kb. They are very similar, with a 5’- methylated cap, a 3’- poly (A) tail (Figure 1) capping the two ends of the remaining open reading frames (ORFs). The genes located in the open reading frame ORF1a express 11 nonstructural proteins (Nsps) from Nsp1 to Nsp11, with the genes of ORF1b expressing proteins from Nsp12 to Nsp16 [8,9,14]. The capping structure at the 5’- end of coronavirus mRNAs plays vital roles in RNA stability, translation initiation and evading host antiviral response [7,15-17]. And at the 3’ end, four or five structural proteins such as Spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins are encoded by other ORFs (Figure 1) [12].
Figure 1: The genome structure of human coronaviruses consists of open reading frames to encode structural proteins and nonstructural proteins. There are 16 nonstructural proteins with different functions. Nsp3 and Nsp5 are a papain-like cysteine protease (PLpro) and 3C like serine protease (3CLpro), respectively. Nsp12 has RNA-dependent RNA polymerase (RdRp) activity, and Nsp13 is a helicase (Hel). Nsp14 and Nsp10/16 have the activities of N7 and 2’-O methyltransferase, respectively. The four structural proteins S, E, M and N are necessary for virus invasion and virion assembly.
The total 16 nonstructural proteins display their different functions. The proteins Nsp3 and Nsp5 are a papain-like cysteine protease (PLpro) and 3C like serine protease (3CLpro), respectively. Protein Nsp12 has RNA-dependent RNA polymerase (RdRp) activity, and Nsp13 is a helicase (Hel), with Nsp14 and Nsp10/Nsp16 having N7 and 2’-O methyltransferase, respectively. The structural proteins S, E, M and N are necessary for virion assembly and virus invasion [9,13]. The S proteins constitute spikes on the surface of the coronavirus and are critical for viruses to enter into the host cells. The E proteins play a critical role in virus assembly and are involved in the pathogenesis of the viruses. The M proteins have three trans-membrane domains that form virions and bind to the nucleocapsid N proteins. The N protein contains two domains that bind to the viral RNA genome through different mechanisms [9,12]. For instance, SARS-CoV has four genes to encode proteins S, E, M and N. HCoV-HKU1 has five genes that express proteins S, E, M, N and HE, respectively. SARS-CoV-2 shares over 96 percent of its RNA genome with other coronaviruses identified before [11,12].
The Infection of Coronaviruses and the Associated Health Problems
MERS-CoV (in 2012), SARS-CoV (in 2002) and SARSCoV-2 (in 2019) are the three key coronaviruses that were originally from animals, eventually infected human and resulted in severe health problems while HCoV-229E, HCoV-NL63, HCoV-OC43 and HCoV- HKU1 are four common human coronaviruses and only cause the common cold or other mild symptoms. The processes of coronavirus infection of cells are very complicated, but are similar. MERS-CoV infects its host’ s respiratory tract via interactions with the DPP4 receptor, activating a series of downstream immune responses [18], as shown with the process of MERS-CoV’s infection in the Figure 2 [19]. The RBD domain of S protein binds to the receptor DPP4 (also called CD26) on the cell membrane. Viruses enter inside cells and release RNA genomes in cytoplasm [20,21]. Viral RNAs bind to ribosomes where to translate viral proteins. The open reading frames of genomes ORF1a and ORF1b were translated into polyprotein 1a (pp1a) and pp1ab, respectively, followed by the cleavage of Papain (PLpro) and 3C-like protease, that produces 16 non-structural proteins. These proteins form transcriptional replication complexes (RTCs) and are encapsulated in double-layered vesicles (DMVs) derived from the endoplasmic reticulum. AU-rich sequences in genomic RNA are called transcriptional regulatory sequences (TRSs). These TRSs are recognized by RTC and transcribed to produce subgenomic RNAs of different lengths, otherwise replication results in a full-length genome. Then the genomic RNA is enveloped and transported to the endoplasmic reticulum-Golgi apparatus by the N protein. The S, M, and E proteins are transported to the rough internal mesh to interact with the N protein to assemble new virus particles. These vesicles then transport matured virus particles to the cell membrane for release outside the membrane. The protein 4a competes with TLR3, RIG-I, and MDA5 and binds double-stranded RNAs (dsRNAs) to evade the host's immune response [22,23].
Figure 2: The life cycle of coronavirus MRES-CoV (Cited from Dr. Durai) (19). First, the S protein of MERS-CoV binds to the receptor DPP4 on the cell membrane, polyprotein 1a (pp1a) and pp1ab followed by Papain (PLpro) and 3C-like protease cleavage then produces 16 nonstructural proteins. These proteins form transcriptional replication complexes (RTCs) and are encapsulated in double-layered vesicles (DMVs) derived from the endoplasmic reticulum. Then, the newly produced genomic RNAs are enveloped and transported to the endoplasmic reticulum-Golgi apparatus by the N protein, and the S, M, and E proteins are transported to the rough internal mesh to interact with the N proteins to assemble new virus particles. These vesicles transport mature virus particles to the cell membrane for release outside the membrane. The double-stranded RNAs (dsRNAs) are produced by genomic RNAs during replication. The 4a protein competes with TLR3, RIG-I, and MDA5 in binding dsRNAs to evade the host's immune responses.
SARS-CoV-2 has similar genomic characteristics to the SARS-CoV that allows it to adapt well to the human respiratory tract. This is primarily attributed to its contact with the angiotensin converting enzyme II (ACE2) ” via its receptor binding domain (RBD) [6]. Although it is worthy to note that SARS-CoV-2 is more contagious, it is estimated to be less pathogenic than SARS-CoV [24]. Some research has already been performed in identifying difference between the two viruses. One initial finding is that SARS-CoV-2 contains a furin-like cleavage site (RRAR) in its Spike (S) protein that is lacking in SARS- CoVs [25]. More studies will be needed to understand how the genomic and structural differences between members of the coronavirus family affect their virulence and pathogenicity.
The Transmission of Coronaviruses
Currently, only seven coronaviruses have been demonstrated for human-to-human transmission. Particularly, the transmission occurs among people via droplets or wastes from coughing, sneezing and other respiratory systems. And three of them, including MERS-CoV, SARS-CoV and SARS-CoV-2 initially pass from animals to human. Current evidence shows that the initial primary transmission of MERS-CoV can be traced to dromedary camels, although bats are thought to be potential intermediate hosts as well [26]. Human-to-human transmission of MERS has been well-documented but has also been shown to be inefficient [27]. With proper implementation of quarantine and infection control protocol, human-to-human transmission of MERS could be greatly curbed [27]. Studies in Saudi Arabia and South Korea has shown that the R0 for MERS to be less than 1 while the incubation period was observed to range from 4-10 days [28].
As previously mentioned, the initial outbreak of SARS-CoV in Southern China quickly spread to nearby Hong Kong, and then on to many other major cities of the world due to Hong Kong’s status as a major travel hub [29]. Beijing, the Chinese capital, became one of the most severely affected megapolises. A rough count in 2004 estimated 66% of cases to have arisen from mainland China and another 22% in Hong Kong, with 4% in Taiwan [30]. The primary hosts of the SARS- CoV is believed to be traced to animals of a reservoir in Southern China, but the virus quickly spread through rapid human-human contact [31]. The SARS-CoV spread through multiple conduits, but the two major routes was transmission through droplets or via contact with infected surfaces [29]. Studies have shown that the SARS-COV could survive in human serum, sputum and feces for over 4 days and even in urine for 3 days while remaining active infectivity. Extermination of the viruses was achieved with >56°C heat and UV irradiation exposure of 90 minutes and 60 minutes, respectively [32].
The mean incubation period of SARS patient is estimated to be 6.4 days with a R0 of around 2.7 [29]. Radiological features of SARS patients shows “ground-glass” opacities that spread rapidly and peaks around days 8-10 after initial onset of the disease [33]. The source of the SARS-CoV-2 outbreak has been successfully traced to the Huanan seafood market in Wuhan, China, where various type of animals were kept in close proximity to each other and to humans. The primary hosts for SARS-CoV-2 are believed to be bats, but human-to-human transmission has quickly developed and is quite rapid, spreading through droplets or other types of close contact [24]. The average incubation period was calculated to be 5.2 days, although in some cases, up to two weeks was reported [34,35]. The WHO estimates of the reproduction number of SARS-CoV-2 ranges from 1.4 to 2.5, but other calculations have posited it to be as high as 3.28 [36].
The Potential Treatments of Coronavirus Infections
Nowadays, there is no any anti-viral vaccine and drug being approved for the treatment of coronavirus infection. Patients infected with the four common coronaviruses (HCoV-229E, HCoV-NL63, HCoV-OC43 and HCoV-HKU1) show mild symptoms such as cold and coughing. However, as for MERS-CoV, SARS-CoV and SARS- CoV-2, patients infected could have severe respiratory problems that can quickly deteriorate into multi-system organ failure and death. There is no effective drug or other treatment available. Currently, patients were mostly treated with supportive care through fluids and nutrition, thus helping their own immune systems to fight off the virus [1,34,37]. SARS patients were often treated with broad-spectrum antibiotics to cover other causes of respiratory disease, before being committed to anti-virals. Ribavirin, for example, was such an anti-viral that was used en-masse in Hong Kong. Low-dose steroids were often given as well [38]. Remdesivir, an antiviral originally designed to treat Ebola, has shown to be effective towards SARS-CoV-2 in-vitro and is currently undergoing extensive medical trials in China [39]. There exists an urgent need to quickly develop anti-CoV vaccines or drugs.
The Development of Coronavirus Vaccines
There are different strategies to develop vaccines including live- attenuated, inactivated, subunits, viral vectors and DNA vaccines [40]. Several vaccines of SARS-CoV and MERS-CoV are under investigation (Table 1). Some vaccines targeting to SARS-CoV and MERS-CoV are currently in different stages of ongoing development, with many of them seeking to target the viral spike (S) protein [41]. As reported, the inactivated SARS-CoV vaccines and the DNA vaccines against S protein can induce strong responses to anti-S antibody and have been tested in Phase I clinical trials. These vaccines all show a potential toxicity such as eosinophilic or other immune-pathological diseases [40,42]. A key domain of the Spike protein that has been explored for the development of SARS-CoV vaccine is the receptor-binding domain (RBD) in the S1 subunit [43]. Kim et al. constructed two adenoviral recombinant vectors (Ad5) encoding the full-length S protein and the S1 extracellular domain of S protein of MERS-CoV, which can induce specific immune responses in mice. However the same responses have yet to be demonstrated in dromedary camels [43].
| Vaccines | Target | Comments | References. |
|---|---|---|---|
| Inactivated SARS-CoV vaccines | Full-length S protein | Induced strong antibody and tested in phase I trials; Induced eosinophilic diseases | (16, 19-21, 23) |
| Live, attenuated SARS-CoV vaccines | E protein | Attenuated E mutant vaccines induce protection in hamsters and mice; Immunocompromised patients have a risk of infection. | (65-68) |
| SARS-CoV DNA vaccines | Full-length S protein | Induce strong antibody and tested in phase I trials; Induce immunopathological diseases. | (16, 21-24) |
| Viral vector vaccines of MERS-CoV | Full-length S protein or S1 extracellular domain of S protein | Induce neutralizing antibody immune responses; Cannot induce T cells responses. | (25, 69) |
| Subunit vaccines of SARS-CoV | RBD of S protein | Induce neutralizing antibodies; May not neutralize SARS-CoV. | -70 |
| DNA vaccines of MERS-CoV | Consensus S protein | Induce protective responses in mice, macaques and camels. | -18 |
Table 1: The SARS-CoV and MERS-CoV vaccines under development.
For MERS-CoV, the vaccines were designed based on recombinant S protein or RBD domain of S protein [42,44,45]. In his work, Dr. Muthumani demonstrated that the DNA vaccine based on the recombinant S protein displayed its potent efficacy in mice, camels and rhesus macaques [45]. Currently, this vaccine is being investigated at phase I clinical trials. During SARS outbreak in 2003, the neutralized antibodies from the memory B cells of SARS survivors or their convalescent sera have been used to treat SARS-CoV patients. In 2012, similar strategies were used to treat MERS-CoV patients during the MERS outbreak [40,46-51].
Vaccines for SARS-CoV-2 are currently under investigation but likely will not become widely available until this episode of the outbreak has ended, much like the case for the SARS-CoV vaccine [52]. Before the vaccine is released, however, the risk of SARS-CoV-2 infection for immune compromised persons, the elderly as well as healthcare workers will remain high in the affected regions. Special considerations should be discussed and put in place to further protect the health of these vulnerable groups.
The Development Of CoV-Targeting Drugs
The drugs for targeting the coronavirus structural proteins
Human coronaviral genomes encode four or five structural proteins including envelope (E), membrane (M), nucleocapsid (N) and spike (S) [53-64]. These structural proteins are necessary for virion assembly and virus invasion and are the targets for drug development. Although there has yet to be any medication approved for specific CoV treatment, scientists will continue their search for anti-CoV drugs. As shown in Table 2, the drug candidates targeting the structural proteins are under investigations. Coronavirus S protein is a 180-200 kDa membrane glycoprotein containing two subunits S1 and S2 that plays a significant role during virus infections [8,11-15]. All human coronaviruses enter into their target cells via the receptor-binding domain (RBD) of S protein [11]. Viral S proteins are thus the critical targets for drug development.
| Targeted structural proteins | Examples | Comments | References |
|---|---|---|---|
| RBD of S protein | m336, m337, m338 and Mersmab1 mAbs for MERS-CoV | High specific binding affinity, but narrow spectrum. | [27, 65] |
| S2 subunit of S protein | HR2P | Inhibit fusion of S protein with DPP4, but narrow spectrum | [28-30] |
| S protein | Griffithsin | Specifically bind to S protein, broad spectrum | [31-33] |
| S, E, M, N | siRNA | Interfere with the expression of SARS-CoV S and E, M. Reduce RNA-binding affinity of N, but narrow spectrum. | [37, 66-69] |
Table 2: The development of drugs targeting for structural proteins in development.
Aiming at MERS-CoV, Dr. Ying et al. developed serial monoclonal antibodies (mAbs) such as m336, m337 and m338. These mAbs specially target the receptor binging domain (RBD) of MERS-CoV S protein and compete with DPP4 for binding to RBD. Especially, the mAb m336 showed its extremely high binding affinity at pM level [54]. Dr. Mori et al identified a novel anti-HIV protein Griffithsin (GRFT) from a red alga from New Zealand [55]. Its anti-viral mechanism was believed to specifically bind to oligosaccharides on HIV gp120. Thus, GRFT might have a broad-spectrum anti-viral activity. Indeed, GRFT was identified not only to inhibit viruses HIV, HCV and JEV, but also to block various coronaviruses including HCoV-229E, HCoV-OC43, SARS-CoV and HCoV-NL63 [56,57]. However, GRFT cannot significantly inhibit the binding interactions of S proteins with its relevant receptors. Small interfering RNA (siRNA), a short double- stranded RNA, also displayed their effects on coronaviruses with the degradation of the target mRNA by complementary base pairing [58].
These siRNAs can be used to silence the crucial genes of coronaviruses and further inhibit the replication of these viruses [59-61]. Peptides can block viruses to interact with the host cells. HR2P, a specific peptide derived from S2 subunit of MERS-CoV S protein, was found to effectively inhibit MERS-CoV fusion and thus block MERS-CoV Infection [62-64].
The drugs for targeting the coronavirus nonstructural proteins
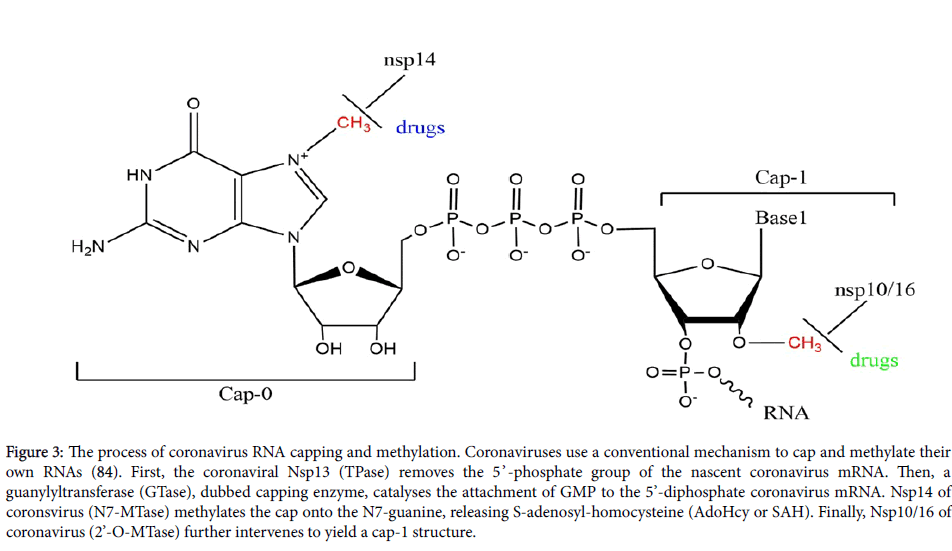
The genes located in the open reading frames ORF1a and ORF1b express 16 nonstructural proteins from Nsp1 to Nsp16 [9]. These nonstructural proteins involved in various viral functions [65,66]. Such proteins, most likely Nsp7 and Nsp16, can direct the synthesis and the process of these coronavirus RNAs. For instance, Nsp3 can induce membrane disordering and regulate proliferation, Nsp3 and Nsp4 together affect viral membrane-paring. Nsp3, Nsp4 and Nsp6 can work together to induce double-membrane vesicles. Nsp14 and Nsp16 regulate mRNA capping, with Nsp12 for RNA-dependent RNA polymerase and Nsp13 for helicase (Figure 3) [10,66,67]. Thus, targeting these nonstructural proteins may provide potential opportunities for anti-coronavirus drug development (Table 3).
Figure 3: The process of coronavirus RNA capping and methylation. Coronaviruses use a conventional mechanism to cap and methylate their own RNAs (84). First, the coronaviral Nsp13 (TPase) removes the 5’ -phosphate group of the nascent coronavirus mRNA. Then, a guanylyltransferase (GTase), dubbed capping enzyme, catalyses the attachment of GMP to the 5’-diphosphate coronavirus mRNA. Nsp14 of coronsvirus (N7-MTase) methylates the cap onto the N7-guanine, releasing S-adenosyl-homocysteine (AdoHcy or SAH). Finally, Nsp10/16 of coronavirus (2’-O-MTase) further intervenes to yield a cap-1 structure.
| Targeted nonstructural proteins | Examples | Comments | References |
|---|---|---|---|
| Nsp3 | E64, Ub-vinylsulfone inhibitor | Specially inhibit DUBs domain of SARS-CoV and HCoV-NL63 PLP2, but not inhibit MERS-CoV | (41, 42) |
| Nsp5 | 3f, 3g and 3m | Inhibit NA and 3CLpro, broad spectrum. | -44 |
| Nsp12 | ATA | Binds the catalytic sites of SARS-CoV’s Nsp12 | -45 |
| Nsp14 | PF35468, PA48202 and PA48523 | Specially inhibit Nsp14 of SARS-CoV, not N7MTase | -58 |
| Nsp10/16 | TP29 | Inhibit SARS-CoV, MERS-CoV, IBV, TGEV, FCoV, MHV Nsp10/16. broad spectrum. | -7 |
Table 3: The strategies to develop drugs via targeting nonstructural proteins.
The nonstructural protein Nsp3 is a papain-like protease that processes polyprotein 1a to release Nsp1, Nsp2 and Nsp3 [68-70]. E64, a cysteine protease inhibitor, can inhibit the Nsp3s of SARS-CoV and HCoV-NL63, but not those of MERS-CoV [71]. PLP2 domains of SARS-CoV and HCoV-NL63 exhibit activity of deubiquitinating enzyme (DUB). Ubiquitin-Vinyl Sulfone (Ub-vinylsulfone, Ub-VS) displayed its potent and specific inhibitory activity against deubiquitinating enzymes (DUBs) [72]. Nsp5 is a 3C-like protease that is vital for coronaviral replication. Thus, it is an attractive target for the development of anti-coronaviral therapeutics [73]. Dr. Kumar et al. [74] reported the pyranones 3f, 3g and 3m could inhibit both neuraminidases (NA) and 3CL protease. All coronaviral Nsp12 proteins have the RNA-dependent RNA polymerase (RdRp) activities responsible for RNA replication [75]. Dr. Yap et al. reported that aurintricarboxylic acid (ATA) that can polymerize and block the interactions of proteins and nucleic acids could inhibit the RdRp activity of SARS-CoV Nsp12 by binding to its catalytic sites [75]. Dr. Sun et al. reported the three natural and microbial product extracts PF35468, PA48202 and PA48523 could inhibit Nsp14 of SARS-CoV [76]. Dr. Ke and his team demonstrated that the two peptides K12 and K29 derived from Nsp10 could inhibit the activity of SARS-CoV’ s nsp16 in a dose-dependent manner, and block the MTase activity of the Nsp10/Nsp16 complex [77]. Another peptide, TP29, inhibited Nsp10 and Nsp16 of six viruses including SARS-CoV, MERS-CoV, IBV, TGEV, FCoV and MHV [7].
Outlook and Challenges
Up to now, a large number of antiviral compounds or prodrugs have been reportedly applied to treat coronaviruses, with some currently under clinical investigation. Meanwhile, there are also many herbal medicines that are being used to treat SARS-CoV-2 patients during the current outbreak in China. However, unfortunately, there has yet to be any formally approved drug made commercially available [78-81]. And another challenge is that, coronaviruses are one of the most multitudinous and rapidly mutated viruses. The novel viruses emerge frequently at unpredictable times. So in the long term, the development of novel, broad-spectrum antiviral drugs may become the ultimate dream for responding to new threats of circulating and emerging CoV infections [82,83].
What is promising is that Remdesivir (GS-5734), an antiviral nucleotide analog, is currently in clinical trials. Remdesivir can significantly target Nsp12 proteins of both SARS-CoV and MERS- CoV, providing an option for the treatment of the novel coronavirus SARS-CoV-2. Although Remdesivir is not yet approved for the market, Gilead and the Chinese Health Bureau have reached an urgent agreement to support and accelerate the phase III clinical trials for Remdesivir to treat the Chinese patients infected with SARS-CoV-2 during the current outbreak. Such a novel anti-coronavirus drug is needed not just in China, but all over the world as Covid-19 continues to spread.
Acknowledgement
We would greatly acknowledge the supports from Shenzhen Science and Technology Program. (Grant No.:KQTD20170810154011370), Xiangtan Institute of Industrial Technology Collaborative Innovation, and Xiangtan Science and Technology Bureau.
References
- Guan W, Xian J (2020) The progress of 2019 Novel Coronavirus (2019-nCoV) event in China. J Med Virol.
- Carlos WG, Dela Cruz CS, Cao B, Pasnick S, Jamil S (2020) Novel Wuhan (2019-nCoV) Coronavirus. Am J Respir Crit Care Med 201:P7-P8.
- Hui DS, E IA, Madani TA, Ntoumi F, Kock R, et al. (2020) The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis 91:264-266.
- Li Q, Guan X, Wu P, Wang X, Zhou L, et al. (2020) Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med
- Wang W, Tang J, Wei F (2020) Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China. J Med Virol 92:441-447.
- Zhu N, Zhang D, Wang W, Li X, Yang B, et al. (2020) A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 382:727-733.
- Wang Y, Sun Y, Wu A, Xu S, Pan R, et al. (2015) Coronavirus nsp10/nsp16 Methyltransferase Can Be Targeted by nsp10-Derived Peptide in vitro and in vivo To Reduce Replication and Pathogenesis. J Virol 89:8416-8427.
- Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY (2016) Coronaviruses: Drug discovery and therapeutic options. Nat Rev Drug Discov 15:327-47.
- Tok T, Tatar G (2017) Structures and Functions of Coronavirus Proteins: Molecular Modeling of Viral Nucleoprotein. International Journal of Virology & Infectious Diseases 2:6.
- Neuman BW (2016) Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles. Antiviral Res 135:97-107.
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature.
- Lu R, Zhao X, Li J, Niu P, Yang B, et al. (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 395:565-574.
- Li B, Si HR, Zhu Y, Yang XL, Anderson DE, et al. (2020) Discovery of Bat Coronaviruses through Surveillance and Probe Capture-Based Next-Generation Sequencing. mSphere 5.
- Su S, Wong G, Shi W, Liu J, Lai AC, et al. (2016) Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol 24:490-502.
- Furuichi Y, Shatkin AJ (2000) Viral and cellular mRNA capping: past and prospects. Adv Virus Res 55:135-184.
- Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, et al. (2010) 2'-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452-456.
- Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, et al. (2011) Ribose 2'-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137-143.
- Mubarak A, Alturaiki W, Hemida MG (2019) Middle East Respiratory Syndrome Coronavirus (MERS-CoV): Infection, Immunological Response, and Vaccine Development. J Immunol Res 2019:6491738.
- Durai P, Batool M, Shah M, Choi S (2015) Middle East respiratory syndrome coronavirus: Transmission, virology and therapeutic targeting to aid in outbreak control. Exp Mol Med 47:e181.
- Du L, Yang Y, Zhou Y, Lu L, Li F, et al. (2017) MERS-CoV spike protein: A key target for antivirals. Expert Opin Ther Targets 21:131-143.
- Kim Y, Cheon S, Min CK, Sohn KM, Kang YJ, et al. (2016) Spread of mutant middle east respiratory syndrome coronavirus with reduced affinity to human CD26 during the South Korean Outbreak. mBio 7:e00019.
- Niemeyer D, Zillinger T, Muth D, Zielecki F, Horvath G, et al. (2013) Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol 87:12489-12495.
- Siu KL, Yeung ML, Kok KH, Yuen KS, Kew C, et al. (2014) Middle east respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J Virol 88:4866-4876.
- Tian HY (2020) [2019-nCoV: new challenges from coronavirus]. Zhonghua Yu Fang Yi Xue Za Zhi 54:E001.
- Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, et al. (2020) The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 176:104742.
- Raj VS, Osterhaus AD, Fouchier RA, Haagmans BL (2014) MERS: Emergence of a novel human coronavirus. Curr Opin Virol 5:58-62.
- Rasmussen SA, Watson AK, Swerdlow DL (2016) Middle East Respiratory Syndrome (MERS). Microbiol Spectr 4.
- Park JE, Jung S, Kim A, Park JE (2018) MERS transmission and risk factors: A systematic review. BMC Public Health 18:574.
- Chan-Yeung M, Xu RH (2003) SARS: epidemiology. Respirology 8 Suppl:S9-14. Cherry JD. 2004. The chronology of the 2002-2003 SARS mini pandemic. Paediatr Respir Rev 5:262-269.
- Vijayanand P, Wilkins E, Woodhead M (2004) Severe acute respiratory syndrome (SARS): A review. Clin Med (Lond) 4:152-160.
- Duan SM, Zhao XS, Wen RF, Huang JJ, Pi GH, et al. (2003) Stability of SARS coronavirus in human specimens and environment and its sensitivity to heating and UV irradiation. Biomed Environ Sci 16:246-255.
- Ooi GC, Daqing M (2003) SARS: Radiological features. Respirology 8 Suppl:S15-9. Special Expert Group for Control of the Epidemic of Novel Coronavirus Pneumonia of the Chinese Preventive Medicine A. 2020. [An update on the epidemiological characteristics of novel coronavirus pneumoniaCOVID-19]. Zhonghua Liu Xing Bing Xue Za Zhi 41:139-144.
- Backer JA, Klinkenberg D, Wallinga J (2020) Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20-28 January 2020. Euro Surveill 25: 2000062.
- Liu Y, Gayle AA, Wilder-Smith A, Rocklov J (2020) The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med.
- Azhar EI, Hui DSC, Memish ZA, Drosten C, Zumla A (2019) The Middle East Respiratory Syndrome (MERS). Infect Dis Clin North Am 33:891-905.
- Tsang K, Zhong NS (2003) SARS: pharmacotherapy. Respirology 8 Suppl:S25-30.
- Wang M, Cao R, Zhang L, Yang X, Liu J, et al. (2020) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30:269-271.
- Perlman S, Vijay R (2016) Middle East Respiratory Syndrome Vaccines. Int J Infect Dis 47:23-28.
- Yong CY, Ong HK, Yeap SK, Ho KL, Tan WS (2019) Recent Advances in the Vaccine Development Against Middle East Respiratory Syndrome-Coronavirus. Front Microbiol 10:1781.
- Du L, Tai W, Zhou Y, Jiang S (2016) Vaccines for the prevention against the threat of MERS-CoV. Expert Rev Vaccines 15:1123-1134.
- Yong CY, Ong HK, Yeap SK, Ho KL, Tan WS (2019) Recent Advances in the Vaccine Development Against Middle East Respiratory Syndrome-Coronavirus. Front Microbiol 10: 1781.
- Du L, Tai W, Zhou Y, Jiang S (2016) Vaccines for the prevention against the threat of MERS-CoV. Expert Rev Vaccines 15: 1123-1134
- Lau YL (2004) SARS: future research and vaccine. Paediatr Respir Rev 5: 300-303.
- Kim E, Okada K, Kenniston T, Raj VS, AlHajri MM, et al. (2014) Immunogenicity of an adenoviral-based Middle East Respiratory Syndrome coronavirus vaccine in BALB/c mice. Vaccine 32: 5975-5982.
- Muthumani K, Falzarano D, Reuschel EL, Tingey C, Flingai S, et al. (2015) A synthetic consensus anti-spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci Transl Med 7: 301ra132.
- Bolles M, Deming D, Long K, Agnihothram S, Whitmore A, et al. (2011) A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J Virol 85: 12201-12215.
- He Y, Zhou Y, Siddiqui P, Jiang S (2004) Inactivated SARS-CoV vaccine elicits high titers of spike protein-specific antibodies that block receptor binding and virus entry. Biochem Biophys Res Commun 325: 445-452.
- Lin JT, Zhang JS, Su N, Xu JG, Wang N, et al. (2019) Safety and immunogenicity from a phase I trial of inactivated severe acute respiratory syndrome coronavirus vaccine. Antivir Ther 12:1107-1113.
- Martin JE, Louder MK, Holman LA, Gordon IJ, Enama ME, et al. (2008) A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a Phase I clinical trial. Vaccine 26: 6338-6343.
- Tseng CT, Sbrana E, Iwata-Yoshikawa N, Newman PC, Garron T, et al. (2012) Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One 7: e35421.
- Yang ZY, Kong WP, Huang Y, Roberts A, Murphy BR, et al. (2004) A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature 428: 561-564.
- Zhang L, Liu Y (2020) Potential Interventions for Novel Coronavirus in China: A Systemic Review. J Med Virol.
- Pyrc K, Berkhout B, van der Hoek L (2007) Antiviral strategies against human coronaviruses. Infect Disord Drug Targets 7: 59-66.
- Ying T, Du L, Ju TW, Prabakaran P, Lau CC, et al. (2014) Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J Virol 88: 7796-7805.
- Mori T, O'Keefe BR, Sowder RC, Bringans S, Gardella R, et al. (2005) Isolation and characterization of griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia sp. J Biol Chem 280: 9345-9353.
- Barton C, Kouokam JC, Lasnik AB, Foreman O, Cambon A, et al. (2014) Activity of and effect of subcutaneous treatment with the broad-spectrum antiviral lectin griffithsin in two laboratory rodent models. Antimicrob Agents Chemother 58: 120-127.
- O'Keefe BR, Giomarelli B, Barnard DL, Shenoy SR, Chan PK, et al. (2010) Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J Virol 84: 2511-2521.
- Gitlin L, Karelsky S, Andino R (2002) Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418: 430-434.
- Wu CJ, Huang HW, Liu CY, Hong CF, Chan YL (2005) Inhibition of SARS-CoV replication by siRNA. Antiviral Res 65: 45-48.
- Channappanavar R, Lu L, Xia S, Du L, Meyerholz DK, et al. (2015) Protective Effect of Intranasal Regimens Containing Peptidic Middle East Respiratory Syndrome Coronavirus Fusion Inhibitor Against MERS-CoV Infection. J Infect Dis 212: 1894-1903.
- Gao J, Lu G, Qi J, Li Y, Wu Y, et al. (2013) Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus. J Virol 87: 13134-13140.
- Lu L, Liu Q, Zhu Y, Chan KH, Qin L, et al. (2014) Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun 5: 3067.
- Snijder EJ, Decroly E, Ziebuhr J (2016) The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv Virus Res 96: 59-126.
- Adedeji AO, Marchand B, Te Velthuis AJ, Snijder EJ, Weiss S, et al. (2012) Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS One 7: e36521.
- Jia Z, Yan L, Ren Z, Wu L, Wang J, et al. (2019) Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res 47: 6538-6550.
- Harcourt BH, Jukneliene D, Kanjanahaluethai A, Bechill J, Severson KM, et al. (2004) Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J Virol 78: 13600-13612.
- Kilianski A, Mielech AM, Deng X, Baker SC (2013) Assessing activity and inhibition of Middle East respiratory syndrome coronavirus papain-like and 3C-like proteases using luciferase-based biosensors. J Virol 87: 11955-11962.
- Yang X, Chen X, Bian G, Tu J, Xing Y, et al. (2014) Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain-like protease. J Gen Virol 95: 614-626.
- Baez-Santos YM, Mielech AM, Deng X, Baker S, Mesecar AD (2014) Catalytic function and substrate specificity of the papain-like protease domain of nsp3 from the Middle East respiratory syndrome coronavirus. J Virol 88: 12511-12527.
- Chen Z, Wang Y, Ratia K, Mesecar AD, Wilkinson KD, et al. (2007) Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J Virol 81: 6007-6018.
- St John SE, Tomar S, Stauffer SR, Mesecar AD (2015) Targeting zoonotic viruses: Structure-based inhibition of the 3C-like protease from bat coronavirus HKU4--The likely reservoir host to the human coronavirus that causes Middle East Respiratory Syndrome (MERS). Bioorg Med Chem 23: 6036-6048.
- Kumar V, Tan KP, Wang YM, Lin SW, Liang PH (2016) Identification, synthesis and evaluation of SARS-CoV and MERS-CoV 3C-like protease inhibitors. Bioorg Med Chem 24: 3035-3042.
- Yap Y, Zhang X, Andonov A, He R (2005) Structural analysis of inhibition mechanisms of aurintricarboxylic acid on SARS-CoV polymerase and other proteins. Comput Biol Chem 29: 212-219.
- Sun Y, Wang Z, Tao J, Wang Y, Wu A, et al. (2014) Yeast-based assays for the high-throughput screening of inhibitors of coronavirus RNA cap guanine-N7-methyltransferase. Antiviral Res 104: 156-164.
- Ke M, Chen Y, Wu A, Sun Y, Su C, et al. (2012) Short peptides derived from the interaction domain of SARS coronavirus nonstructural protein nsp10 can suppress the 2'-O-methyltransferase activity of nsp10/nsp16 complex. Virus Res 167: 322-328.
- Agrawal AS, Garron T, Tao X, Peng BH, Wakamiya M, et al. (2015) Generation of a transgenic mouse model of Middle East respiratory syndrome coronavirus infection and disease. J Virol 89: 3659-3670.
- McCray PB, Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, et al. (2007) Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol 81: 813-821.
- Roberts A, Deming D, Paddock CD, Cheng A, Yount B, et al. (2007) A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog 3: e5.
- Tseng CT, Huang C, Newman P, Wang N, Narayanan K, et al. (2007) Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. J Virol 81: 1162-1173.
- Brown AJ, Won JJ, Graham RL, Dinnon KH, Sims AC, et al. (2019) Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res 169: 104541.
- Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, et al. (2017). Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med 9: eaal3653.
Citation: Wang S, Zhang Y, Liu S, Peng H, Mackey V, et al. (2020) Coronaviruses and the Associated Potential Therapeutics for the ViralInfections. J Infect Dis Ther 8: 417. DOI: 10.4172/2332-0877.1000417
Copyright: © 2020 Wang S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 3555
- [From(publication date): 0-2020 - Nov 15, 2025]
- Breakdown by view type
- HTML page views: 2670
- PDF downloads: 885