Computational Analysis of Cholesterol Binding and Pore-Lining Regions in Alpha-Synuclein: Role in Mitochondrial Function
Received: 12-Sep-2017 / Accepted Date: 19-Sep-2017 / Published Date: 26-Sep-2017 DOI: 10.4172/2161-0460.1000378
Abstract
Alpha-synuclein (α-syn) protein is the major component of Lewy bodies, which are characteristic pathological trademarks for neurodegenerative diseases (e.g. Parkinson’s and Alzheimer’s diseases). It is primarily expressed in neural tissue, with smaller amounts found in heart, muscle and other tissues. The canonical form found in Homo sapiens (α-syn-1) contains 140 residues and interacts with neuronal mitochondria via an N-terminal 32 residue mitochondrial-targeting signal. All isoforms (there are 3) have multiple highly conserved lipid binding (KTKE(Q)G(Q) V) motifs, thought to mediate binding to phospholipid membranes. Two isoforms also contain an EF-hand-like (helixloop- helix) sequence found in a large family of calcium-binding proteins, as well as three copper binding sites. We investigate protein topology using computational analysis and find that each isoform contains a pore-lining region, two cholesterol-binding (CRAC/CARC) and three or four lipid binding motifs, with one cholesterol motif overlapping the pore-lining region. Two lipid-binding motifs also overlap the N-terminal mitochondrial-targeting region consistent with evidence that α-syn inserts into mitochondrial inner membrane. α-Syn-1 reportedly occurs physiologically as a helically folded tetramer that requires N-terminal acetylation. Thus, each α-syn-1 tetramer could contain 4 mitochondrial targeting regions, up to 4 pore-lining regions, 4 EF-hand domains, 8 bound cholesterol molecules and 16 lipid binding motifs with pore-lining regions merging to form a membrane channel. Cholesterol binding to CRAC motifs may in turn facilitate protein folding, Ca2+-channel formation, as well as mitochondrial membrane lipid-protein interactions, altering mitochondrial bioenergetics. Disruption of mitochondrial bioenergetics may be involved in the pathogenesis of Alzheimer’s disease and Parkinsonism.
Keywords: Alpha-synuclein; Pore-lining regions; Mitochondrial targeting regions; Cholesterol-binding (CRAC/CARC) motifs; KTKEGV motifs; EF-hand domains; Copper binding sites
Introduction
Alpha-synuclein (α-syn) is a small (14 kDa), acidic protein that is highly conserved in vertebrates, making up as much as 1% of all proteins in the cytosol of brain cells [1]. The α-Syn protein is the major component of Lewy bodies, which are characteristic pathological trademarks for neurodegenerative diseases (e.g. Parkinson’s and Alzheimer’s diseases; rev. [2]). It is predominantly expressed in the neocortex, hippocampus, substantia nigra, thalamus and cerebellum as a neuronal protein, and can also be found in the neuroglial cells. It is also expressed in low concentrations in all tissues examined except liver [see UniprotKB-P37840 (SYUA_HUMAN) for Gene expression databases]. α-Syn is localized in the presynaptic termini in both free and membrane-bound forms, with about 15% of α-syn being estimated as membrane-bound at any given moment in neurons [3]. α-Syn is also highly expressed in the mitochondria in the olfactory bulb, hippocampus, striatum and thalamus [4,5]. Cerebral cortex and cerebellum are exceptions, with each containing high levels of cytosolic α-syn but very low mitochondrial levels [4]. Devi et al. [6] have shown that α-syn contains a 32 amino acid mitochondrial targeting signal with evidence that accumulation of α-syn in human dopaminergic neurons caused reduced mitochondrial complex I activity and increased production of reactive oxygen species. Complex I (NADH:ubiquinone) is the largest multimeric enzyme complex of the mitochondrial respiratory chain, responsible for electron transport and the generation of a proton gradient across the mitochondrial inner membrane to drive ATP production. A number of proteins have been identified as assembly factors for complex I biogenesis and numerous mitochondrial diseases have been associated with complex I dysfunction (rev [7]).
In the normal brain, α-syn is found at the tips of neurons in specialized structures called presynaptic terminals [1]. Within these structures, α-syn interacts with phospholipids [8] and other neuronal proteins [3]. A fraction of α-syn interacts with mitochondria, and increased α-syn can produce mitochondrial fragmentation and impair mitochondrial complex 1 activity [9]. Other studies suggest that α-syn plays a role in maintaining a supply of synaptic vesicles in presynaptic terminals by synaptic vesicle clustering [10]. α-Syn reportedly occurs physiologically as a helically folded tetramer that resists aggregation [11]. The usual form of the protein, and the one most investigated, is the full-length protein of 140 amino acids. Other isoforms are alphasynuclein- 3, which lacks residues 41-54 due to loss of exon 3; and alphasynuclein- 2 [12], which lacks residue 103-130 due to loss of exon 5 [13].
Here we identify additional protein domains and moieties in the three α-syn isoforms, and analyze the role of protein topology in α-syn-1 physiology. A protein domain is a conserved part of a given protein sequence and (tertiary) structure that can evolve, function, and exist independently of the rest of the protein chain. Each domain forms a compact three-dimensional structure and often can be independently stable and folded and responsible for a particular function or interaction, contributing to the overall role of a protein (e.g. [14]). A super secondary structure such as a helix-turn-helix which appears in a variety of proteins is often defined as a motif and may describe the connectivity between secondary structural elements [15]. As described here, α-syn contains multiple protein domains including a 32 residue N-terminal mitochondrion-targeting sequence, a pore-lining region, multiple cholesterol binding (CRAC/CARC) motifs and lipid-binding KTKE(Q)G(Q)V motifs, an EF-hand motif and several copper binding sites with cholesterol binding motifs overlapping specific domains and super secondary structures. Based on protein topology analysis, we find that each α-syn tetramer may contain 4 pore-lining regions that form channels and up to 8 bound cholesterol molecules overlapping other motifs and serve to modulate ion movement and interaction with mitochondrial membranes.
Materials and Methods
Sequence analysis of α-syn isoforms
The amino acid sequences of Homo sapiens α-syn isoform 1 (Accession #P37840-1), α-syn isoform 2 (Accession #P37840-2) and α-syn isoform 3 (Accession #P37840-3) were downloaded from the ExPASy Proteomics Server of the Swiss Institute of Bioinfomatics (http://www.expasy.org; http://www.uniprot.org). Amino acid sequences were compared using the Pairwise Sequence Alignment software (LALIGN) at http://www.ebi.ac.uk/Tools/services/web lalign/ to find internal duplications by calculating non-intersecting local alignments. The Emboss Water protocol (version 36.3.5e Nov, 2012; preload8) used here employs the Smith-Waterman algorithm (with modified enhancements) to calculate the local alignment of two sequences.
Computational analysis of α-syn isoforms
Secondary structure, transmembrane helix and/or pore-lining region predictions were determined by computational analysis. PSIPRED is a simple and accurate secondary structure prediction method, incorporating two feed-forward neural networks which produce an output obtained from Position Specific Iterated - Blast. PSIPRED 3.2 achieves an average Q3 score of 81.6% [16] and is available at http://bioinf.cs.ucl.ac.uk/psipred/. Transmembrane helices and pore-lining regions in transmembrane protein sequences in this study were predicted using the method of Nugent and Jones [17]. These workers have trained [17] a support vector machine classifier to predict the likelihood of a TM helix being involved in pore formation. This approach has a prediction accuracy of 72%, while a support vector regression model is able to predict the number of subunits participating in the pore with 62% accuracy.
Analysis of cholesterol (CRAC, CARC) motifs
CRAC is a short linear amino acid motif that mediates binding to cholesterol and stands for Cholesterol Recognition/Interaction Amino acid Consensus sequence [18]. In a N-terminus to C-terminus direction the motif consists of a branched apolar Leu (L), Ileu (I) or Val (V) residue, followed by a segment containing 1-5 of any residues, followed by an aromatic Tyr (Y)/Phe (F) residue, a segment containing 1-5 of any residues, and finally a basic Lys (K) or Arg (R). In the one letter amino acid codes the algorithm is (I/L/V) – X1-5 – (Y/F) – X1-5- (K/R). A second cholesterol recognition domain similar to the CRAC domain (CARC) has been identified (cit. [18]) but exhibits the opposite orientation along the polypeptide chain (“inverted CRAC”), i.e., (K/R) – X1-5 – (Y/F) – X1-5 – (L/V).
EF-hand domain analysis
The EF-hand is a helix-loop-helix structural domain or motif found in a large family of calcium-binding proteins [19]. The EF-hand motif contains a helix-loop-helix topology, much like the spread thumb and forefinger of the human hand, in which the Ca2+ ions are coordinated by ligands within the loop. The EF-hand consists of two alpha helices linked by a short loop region (usually about 12 amino acids) that usually binds calcium ions.
11-Residue lipid-binding domains
An antipathic N-terminal region of α-syn (residues 1-63) dominated by four 11-residue repeats including the consensus sequence KTKE(Q)G(Q)V has a structural alpha helix propensity similar to apolipoprotein-binding domains (e.g. [20]). Similarities in these repeats with those found in apolipoproteins have suggested a possible role in the association of α-syn with lipids through the formation of an α-helical secondary structure [21]. When mixed with lipid vesicles, including negatively charged phospholipids, the N-terminal region of α-syn adopts a α-helical conformation [22].
Results and Discussion
Analysis of alpha-synuclein (α-syn) protein topology in terms of pore-lining regions, disordered regions and Dompred and DomSSEA boundaries
Nugent and Jones [17] have developed a method (MEMSATSVM) to predict pore-lining regions in transmembrane (TM) proteins and to distinguish pore-lining regions from TM helices. In predicting pore stoichiometry, four features were used to train a support vector machine (SVM) model: sequence length, the number of pore lining residues, topology and the number of pore-lining helices, with the target value set to the number of subunits contracting the pore within the membrane region. Pore-lining regions are usually enriched in negatively (e.g. E, D) or positively charged residues (e.g. K, R, H). Using this approach, Nugent et al. 16] were able to identify pore-lining regions and to predict both the likelihood of transmembrane helices involved in pore (channel) formation and to determine the number of subunits required to form a complete pore or ion channel.
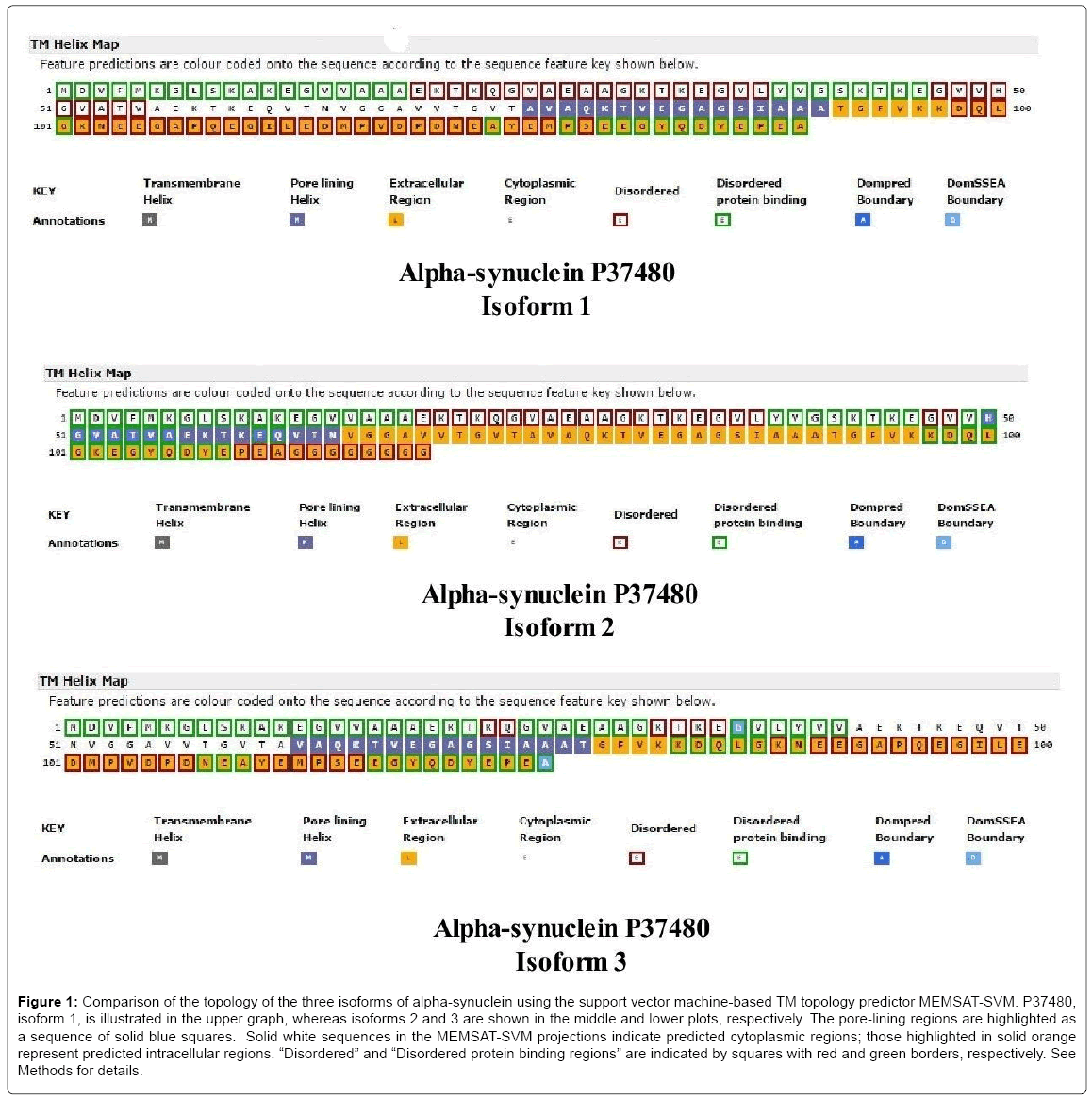
Including the initiator methionine, the α-syn isoforms vary from 140 residues (isoform-1) to 112 residues (isoform-2) and 126 residues (isoform-3). The pore-lining regions, disordered protein binding regions, as well as Dompred and DomSSEA boundaries are compared (Figure 1) in the three α-syn isoforms of Homo sapiens produced by alternative splicing (P37840-1, top; P37840-2, middle; P37840-3, bottom). P37840-1 has been chosen as the ‘canonical’ sequence (also known as NACP140). The solid blue squares indicate pore-lining regions whereas the extracellular region is indicated as orange-filled squares with the predicted intracellular region shown as colorless squares (Figure 1). As indicated by the key annotations (lower part of each plot), squares with dark red and green rims, respectively, represent disordered and disordered protein binding regions. As shown, each isoform contains a 16-residue transmembrane pore-lining region which should facilitate insertion of α-syn into mitochondrial membranes. The pore-lining region of the canonical form (76A-A91) is located within domain 2 (residues 61-95) of the three domains (1-60; 61-95; 96-140) identified in α-syn. The three isoforms are similar in that all C-terminal regions are predicted to be extramitochondrial (or “extracellular”) but differ in their disordered regions. Using LALIGN, a comparison of isoforms 1 and 2 indicates a Waterman-Eggert score of 616, with 80.0% identity (80.0% similar) in 140 amino acid overlaps (1-140:1-112). Residues 103 to 130 are deleted in isoform 2. In comparison, isoforms 1 and 3 exhibit a score of 752, with 90.0% identity (90.0% similar) in 140 amino acid overlaps (1-140:1-126). Residues 41 to 55 are deleted in isoform 3. A major difference in topology is a lateral shift of the porelining region in each isoform.
Figure 1: Comparison of the topology of the three isoforms of alpha-synuclein using the support vector machine-based TM topology predictor MEMSAT-SVM. P37480, isoform 1, is illustrated in the upper graph, whereas isoforms 2 and 3 are shown in the middle and lower plots, respectively. The pore-lining regions are highlighted as a sequence of solid blue squares. Solid white sequences in the MEMSAT-SVM projections indicate predicted cytoplasmic regions; those highlighted in solid orange represent predicted intracellular regions. “Disordered” and “Disordered protein binding regions” are indicated by squares with red and green borders, respectively. See Methods for details.
Distribution of cholesterol-binding (CRAC/ CARC) domains, KTKE(Q)G(Q)V repeat motifs and copper binding sites and C-terminal EF-hand-like domains
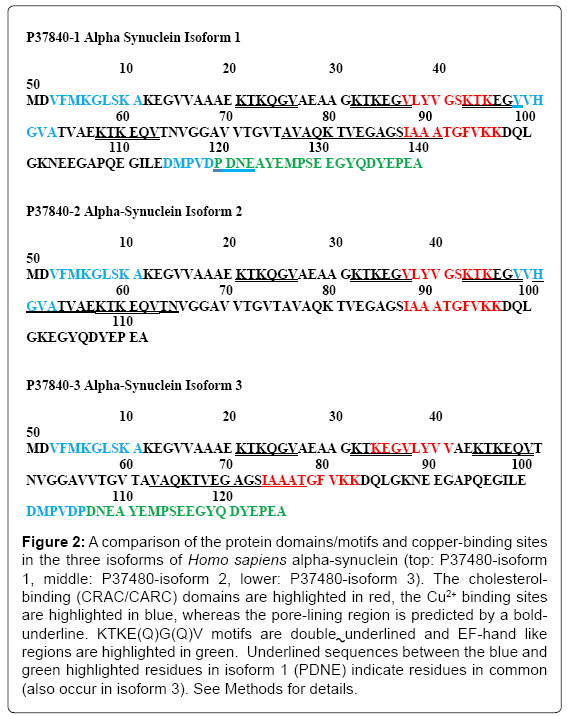
Figure 2 compares the pore-lining regions (sequences are bold underlined), cholesterol binding (CRAC/CARC) domains (highlighted in red), KTKE(Q)G(Q)V motifs (double underlined) and EF-hand like regions (green highlighted) in the α-syn isoforms. The differential copper binding regions are highlighted in blue (cf. [23]). Each α-syn isoform contains two cholesterol binding (CRAC or CARC) motifs, one of which overlaps a pore-lining region and/or a KTKE(Q)G(Q)V repeat motif in all 3 isoforms. Recent studies have defined new roles for copper metabolism in cell proliferation, signaling and disease including SCO1 and SCO2 that are essential for the assembly of the catalytic core of cytochrome c oxidase (rev. [24]). Miotto et al. [25] have shown that N-terminal acetylation facilitates Cu+ binding to the naturally occurring form of α-syn at Met-1 and Met-5 and to a lesser extent to Met-116, -127 and His-50. More recent studies by Ranjan et al. [23] report that Cu2+ first binds to residues in the stretch of amino acids 48-53 with a higher affinity, which is followed by the binding at residues 3-11 and residues 115-123, respectively, in the case of the canonical form of alpha-syn. Isoforms 1 and 3 (but not isoform 2) contain an EF-handlike (helix-loop-helix) C-terminal sequence found in a large family of calcium-binding proteins. As predicted by the MEMSAT-SVM algorithm in (Figure 1), the EF-hand domain is extra mitochondrial (or extracellular), in a position to modulate Ca2+ binding to the α-syn at the mitochondrial surface.
The CRAC sequence has been suggested to relate to the propensity of membrane proteins to be incorporated into cholesterol-rich lipid domains as well as promote segregation of cholesterol into lipid domains [18]. It is important to note a possibility of “false positive” results in CRAC/CARC motifs. Many proteins are shown to have multiple CRAC/CARC motifs but have not been confirmed as actual cholesterol binding domains. Modeling studies by Fantini et al. [26] indicate that a cholesterol molecule can bind to the CRAC domain with energy of interaction of -44.4 kJ mol-1 whereas the CARC domain binds with a significantly higher energy of interaction of -62.4 kJ mol-1. The presence of a CARC or CRAC motif in a protein usually indicates its intrinsic capability to interact with cholesterol, provided that the motifs are accessible to cholesterol molecules. Based on energy considerations, protein folding can be affected by cholesterol binding and protein folding may be different in the presence and absence of cholesterol. It may be possible to characterize cholesterol binding using 1H/13C-NMR with purified preparations of α-syn. It should also be noted that methionine is the N-terminal amino acid in each of the α-syn isoforms (Figure 2). In a biological system, protein synthesis is initiated universally with the amino acid methionine (cit. [27]) which is subsequently removed by enzymes in the cell. The codon initiator methionine is therefore listed in protein data files, although it may not be present physiologically. A contribution from N-terminal methionine to protein topology/interactions has not been ruled out.
Figure 2: A comparison of the protein domains/motifs and copper-binding sites in the three isoforms of Homo sapiens alpha-synuclein (top: P37480-isoform 1, middle: P37480-isoform 2, lower: P37480-isoform 3). The cholesterolbinding (CRAC/CARC) domains are highlighted in red, the Cu2+ binding sites are highlighted in blue, whereas the pore-lining region is predicted by a boldunderline. KTKE(Q)G(Q)V motifs are double underlined and EF-hand like regions are highlighted in green. Underlined sequences between the blue and green highlighted residues in isoform 1 (PDNE) indicate residues in common (also occur in isoform 3). See Methods for details.
Distribution of mitochondrial-targeting sequences and C-terminal EF-hand-like domains
The intracellular sorting of newly synthesized precursor proteins (preproteins) to mitochondria depends on the “mitochondria-targeting sequence” (MTS) which is located at the N-termini of the preproteins (rev. [28]). MTS is required not only for targeting newly synthesized preproteins to mitochondria, but also for several steps along the mitochondrial protein import pathway. MTS is described as a multirole sorting which specifically interacts with various components along the mitochondrial protein import pathway. Targeting of proteins to different cellular locations is often mediated by N-terminal “topogenic sequences” (rev. [29]). However, most mitochondrial proteins are encoded by the nuclear genome and thus have to be imported into mitochondria from the cytosol (rev. [30]). In the case of α-syn, the N-terminal 32 amino acids have been identified as an MTS [6]. As can be seen in Figure 2, the 32 amino acids are identical in all three isoforms, with all three containing a copper binding site (VFMKGLSKA) and a lipid-binding KTKE(Q)G(Q)V motif.
As also shown in Figure 2, the last 20 residues of α-syn represent the EF-hand domain. Based on protein topology, this may exist as a random coil structure due to its low hydrophobicity and high net negative charge. Trexler and Rhoades [31] have demonstrated that N-terminal acetylation is critical for forming α-helical oligomers of α-syn. Further evidence for the tetrameric conformation has been obtained by Bartels et al. [32], Pochapsky [33] and Dettmer et al. [34]. As noted by Trexler and Rhoades [31], acetylation removes the N-terminal charge, making this region more hydrophobic. Increased hydrophobicity would favor protein-protein interactions of α-syn involving hydrophobic packing, either in folding to a native tetramer or in aggregations found in Parkinson’s disease. It should also be emphasized that studies using an E. coli expression system to prepare large amounts of α-syn from human C-DNA (e.g. [35] would generate non-acetylated protein monomers. Acetylated α-syn can be produced by co-transfecting E. coli with both α-syn and N-terminal acetylated B complex plasmids [11]. The pattern emerging from numerous studies suggests that the N-acetylated tetrameric helix-rich form is important to α-syn physiology.
Amphipathic α-helical domain binding to phospholipid membranes: Role of conserved KTKEGV heteromeric motifs and formation of helically folded tetramers
Amphipathic α-helical domains in the N-terminus of α-syn reportedly mediate binding to phospholipid membranes [36]. This domain is structurally similar to the class A2 lipid-binding motif described for the exchangeable apolipoproteins. As proposed by George and Yang [36], α-syn is unstructured in aqueous solution, but shifts to a unique α11/3-helical conformation in the presence of phospholipid vesicles or SDS. In addition, α-syn binds fatty acids, particularly those with long polyunsaturated acyl chains (PUFAs), and these interactions can induce irreversible multimerization of the protein. The N-terminal domain (residues 1-70) of the canonical α-syn-1 contains a positively charged region that includes the 11-amino acid heteromeric repeats. Each 11-amino acid repeat contains a highly conserved KTKE(Q) G(Q)V hexameric motif that is also present in the α-helical domain of apolipoproteins (rev. [36]. The core region of α-syn (residues 61- 95, also known as NAC) is involved in fibril formation and, as shown in (Figure 2), includes the pore-lining region and a CRAC motif. The C-terminal region of α-syn (residues 96-140) is defined as an “acidic tail” of 43-amino acid residues, containing 10 Glu and 5 Asp residues, and contains the EF-hand motif.
Conclusion
We find that current protein computational methods indicate that α-syn contains previously unidentified protein domains and motifs. These include a pore-lining region (Figure 1) as well as two cholesterol-binding (CRAC) motifs per α-syn molecule (Figure 2). The α-syn protein domains and motifs in isoform 1 identified to date are summarized in (Table 1). All α-syn isomers contain N-terminal 32 residue mitochondrial-targeting signal and coupled with the finding that α-syn is highly expressed in the mitochondria in the olfactory bulb, hippocampus, striatum and thalamus [4] indicates that α-syn is an important mitochondrial protein. Other evidence in the literature indicates that, under physiological conditions, the α-syn molecule is acetylated in the N-terminal region and forms tetramers (e.g. [11]).
| Protein Domains/Motifs | Amino Acid Sequence | Function |
|---|---|---|
| Mitochondria-Targeting Signal | 1-32 | Directs to Mitochondrial Membrane |
| Pore-Lining Region | 76-91 | Membrane Channel-Forming |
| CRAC Motifs | 37-45; 88-97 | Cholesterol Binding Sites |
| “KTKE(Q)G(Q)V” Motifs | 21-26; 32-37; 43-48; 58-63 | Phospholipid-Binding Sequence |
| EF-Hand-Like Sequence | 120-140 | Ca2+ Binding Helix-Turn-Helix |
| Cu2+ Binding Sites | 3-11; 48-53; 115-123 | Assembly of Catalytic Core of Cytochrome C Oxidase |
Table 1: Summary of protein domains, motifs and specific binding sites in Homo sapiens α-Synuclein (Isoform 1).
The mitochondrial-targeting sequence in the N-terminal region indicates that α-syn may selectively insert in the inner mitochondrial membranes of neural cells [6,9,37]. Thus, the pore-lining regions within tetramers may merge to form a mitochondrial membrane ion channel and alter mitochondrial bioenergetics. All α-syn isoforms also contain multiple highly conserved phospholipid-binding motifs (KTKE(Q) G(Q)V), thought to mediate binding to phospholipid membranes (e.g. [34]), which in the tetramer would involve up to 16 phospholipid binding (KTKE(Q)G(Q)V) motifs. Cholesterol-containing pore-lining regions are known to form Ca2+ ion channels [18,26] which together with the EF-hand in the C-terminal region of the molecule would regulate Ca2+ movement across the mitochondrial membrane. It has been proposed that memory reconsolidation and its maintenance depend both on voltage-dependent Ca2+ channels as well as calcium/calmodulindependent protein kinase in the hippocampus [38]. Ion channel formation in plasma/mitochondrial membranes can be facilitated by exercise and account for the increase in both brain cognition and the beneficial effects of physical activity on selective aspects of brain function (cit. [39]). An excess of α-syn (e.g. in Parkinson’s disease) may produce mitochondrial calcium overload, resulting in mitochondrial dysfunction/destruction and neuronal cell death [40].
Acknowledgement
This research was supported in part by National Institutes of Health research grants HD10463 and GM-071324.
References
- Beyer K, Ariza A (2013) α-Synuclein posttranslational modification and alternative splicing as a trigger for neurodegeneration. Mol Neurobiol 47: 509-524.
- McLean PJ, Kawamata H, Ribich S, Hyman BT (2000) Membrane association and protein conformation of alpha-synuclein in intact neurons. Effect of Parkinson's disease-linked mutations. J Biol Chem 275: 8812-8816.
- Liu G, Zhang C, Yin J, Li X, Cheng F, et al. (2009) Alpha-synuclein is differentially expressed in mitochondria from different rat brain regions and dose-dependently down-regulates complex I activity. Neurosci Lett 454: 187-192.
- Zhang L, Zhang C, Zhu Y, Cai Q, Chan P, et al. (2008) Semi-quantitative analysis of alpha-synuclein in subcellular pools of rat brain neurons: An immunogold electron microscopic study using a C-terminal specific monoclonal antibody.Brain Res 1244: 40-52.
- Devi L, Raghavendran V, Prabhu BM, Avadhani NG, Anandatheerthavarada HK (2008) Mitochondrial import and accumulation of a-synuclein impair complex 1 in human dopaminergic neuronal cultures and Parkinson disease brain. J Biol Chem 283: 9089-9100.
- Mimaki M, Wang X, Mckenzie M, Thorburn DR, Ryan MT (2012) Understanding mitochondrial complex I assembly in health and disease. Biochim Biophys Acta 1817: 851-862.
- Zigoneanu IG, Yang YJ, Krois AS, Haque E, Pielak GJ (2012) Interaction of α-synuclein with vesicles that mimic mitochondrial membranes. Biochim Biophys Acta 1818: 512-519.
- Nakamura K (2013) α-Synclein and mitochondria: Partners in crime? Neurotherapeutics 10: 391-399.
- Diao J, Burre J, Vivona S, Cipriano DJ, Sharma M, et al. (2013) Native α-synuclein induces clustering of synaptic-vesicle mimics via binding to phosoholipids and synaptobrevin-2/VAMP2. Elife 2: e00592.
- Bartels T, Choi IG, Selkoe DJ (2011) α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature 477: 107-110.
- Uéda K, Saitoh T, Mori H (1994) Tissue-dependent alternative splicing of mRNA for NACP, the precursor of non-A beta component of Alzheimer's disease amyloid.Biochem Biophys Res Commun 205: 1366-1372.
- Beyer K (2006) Alpha-synuclein structure, posttranslational modification and alternative splicing as aggregation enhancers. Acta Neuropathol 112: 237-251.
- Chothia C, Lesk AM, Gherardi E, Tomlinson IM, Walter G, et al. (1992) Structural repertoire of the human VH segments. J Mol Biol 227: 799-817.
- Chiang YS, Gelfand TL, Kaster AE, Gelfand IM (2007) New classification of super secondary structures of sandwich-like proteins uncovers strict patterns of strand assemblage. Proteins 68: 915-921.
- Nugent T, Ward S, Jones DT (2011) The MEMPACK alpha-helical transmembrane protein structure prediction server. Bioinformatics 27: 1438-1439.
- Nugent T, Jones DT (2012) Detecting pore-lining regions in transmembrane protein sequences. BMC Bioinformatics 13: 169.
- Fantini J, Barrantes FJ (2013) How cholesterol interacts with membrane proteins: An exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front Physiol 4: 31.
- Nelson MR, Thulin E, Fagan PA, Forsén S, Chazin WJ (2002) The EF-hand domain: A globally cooperative structural unit. Protein Sci 11: 198-205.
- Davidson WS, Jonas A, Clayton DF, George JM (1998) Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J Biol Chem 273: 9443-9449.
- Fortin DL, Troyer MD, Nakamura K, Kubo S, Anthony MD, et al. (2004) Lipid rafts mediate the synaptic localization of alpha-synuclein. J Neurosci 24: 6715-6723.
- Ulmer TS, Bax A, Cole NB, Nussbaum RL (2005) Structure and dynamics of micelle-bound human alpha-synuclein. J Biol Chem 280: 9595-9603.
- Ranjan P, Ghosh D, Yarramala DS, Das S, Maji SK, et al. (2017) Differential copper binding to alpha-synuclein and its disease-associated mutants affect the aggregation and amyloid formation. Biochim Biophys Acta 1861: 365-374.
- Turski ML, Thiele DJ (2009) New roles for copper metabolism in cell proliferation, signaling and disease. J Biol Chem 284: 717-721.
- Miotto MC, Valiente-Gabioud AA, Rossetti G, Zweckstetter M, Carloni P, et al. (2015) Copper binding to the N-terminally acetylated, naturally occurring form of alpha-synuclein induces local helical folding. J Am Chem Soc 137: 6444-6447.
- Fantini J, Di Scala C, Evans LS, Williamson PT, Barrantes FJ (2016) A mirror code for protein-cholesterol interactions in the two leaflets of biological membranes. Sci Rep 6: 21907.
- Drabkin HJ, RajBhandary UL (1998) Initiation of protein synthesis in mammalian cells with codons other than AUG and amino acids other than methionine. Mol Cell Biol 18: 5140-5147.
- Omura T (1998) Mitochondria-targeting sequences, a multi-role sorting sequence recognized at all steps of protein import into mitochondria. J Biochem 123: 1010-1016.
- Fluman N, Tobiasson V, von Heijne G (2017) Stable membrane orientations of small dual-topology membrane proteins. Proc Natl Acad Sci USA 114: 7987-7992.
- Paschen SA, Neupert W (2001) Protein import into mitochondria. IUBMB Life 52: 101-112.
- Trexler AJ, Rhoades E (2012) N-Terminal acetylation is critical for forming α-helical oligomer of α-synuclein. Protein Sci 21: 601-605.
- Bartels T, Kim C, Luth ES, Selkoe DJ (2014) N-alpha-acetylation of a-synuclein increases its helical folding propensity, GM1 binding specificity and resistance to aggregation. PLoS ONE 9: e103727.
- Pochapsky TC (2015) From intrinsically disordered protein to context-dependent folding: The a-synuclein tetramer is teased out of hiding. Proc Natl Acad Sci USA 112: 9502-9503.
- Dettmer U, Newman AJ, von Saucken VE, Bartels T, Selkoe D (2015) KTKEGV repeat motifs are key mediators of normal a-synuclein tetramerization: Their mutation causes excess monomers and neurotoxicity. Proc Natl Acad Sci USA 112: 9596-601.
- Yonemoto IT, Wood MR, Batch WE, Kelly JW (2009) A general strategy for the bacterial expression of amyloidogenic peptides using BCL-XL-1/2 fusions. Protein Sci 18: 1978-1986.
- George JM, Yang ML (2013) a-Synuclein physiology and membrane binding. Madam Curie Bioscience Database [Internet]. Austin (TX): Landes Biosciences.
- Di Malo R, Barrett PJ, Hoffman EK, Barrett CW, Zharikov A, et al. (2016) α-Synuclein binds to TOM20 and inhibits mitochondrial protein import in Parkinson’s disease. Sci Transl Med 8: 342ra78.
- Da Silva WC, Cardosa G, Bonini JS, Benetti F, Izquierdo I (2013) Memory consolidation and its maintenance depend on l-voltage-dependent calcium channels and CaMKii functions regulating protein turnover in the hippocampus. Proc Natl Acad Sci USA 110: 6566-70.
- Hillman CH, Erickson KI, Kramer AF (2008) Be smart, exercise your heart: Exercise effects on brain and cognition. Nat Rev Neurosci 9: 58-65.
- Stefanis L (2012) α-Synuclein in Parkinson's DiseaseCold Spring Harb Perspect Med 2: a009399.
Citation: Morrill GA, Kostellow AB, Gupta RK (2017) Computational Analysis of Cholesterol Binding and Pore-Lining Regions in Alpha-Synuclein: Role in Mitochondrial Function. J Alzheimers Dis Parkinsonism 7: 378. DOI: 10.4172/2161-0460.1000378
Copyright: © 2017 Morrill GA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 5276
- [From(publication date): 0-2017 - Apr 02, 2025]
- Breakdown by view type
- HTML page views: 4421
- PDF downloads: 855