Comparison of 3 Tests for Plasma HIV-1 RNA Quantitation of Non-B Subtypes in Patients Infected with HIV-1 in N’Djamena-Chad: Cobas AmpliPrep/Cobas TaqMan HIV-1 Test Version 2.0, Abbott m2000 RealTime and Generic HIV Viral Load® Assays
Received: 21-Aug-2018 / Accepted Date: 27-Sep-2018 / Published Date: 02-Oct-2018 DOI: 10.4172/2332-0877.1000382
Keywords: Plasma viral load; Cobas ampliprep/cobas taqMan; Abbott real-time; Generic HIV viral load® ; Resource-limited countries; HIV-1
Introduction
Antiretroviral Therapy (ART), regardless of the first line or subsequent lines, including after multiple failures, is administered to the patient with the objective to obtain and maintain undetectable plasma Viral Load (VL) [1]. Measurements of VL and resistance tests are used as markers of treatment efficacy. But for their usefulness, these methods require adequate equipment and qualified personnel. The complexity of these techniques and their high cost means that they are not used or at least rarely in Resource-Limited Countries (RLCs). Although there are patients’ follow-up structures such as hospitals and laboratories, most are limited and under-equipped to address the problem of diagnosis and monitoring of HIV infection. In addition, logistical difficulties such as the transport of blood samples and respect for the cold chain make it difficult to monitor patients living in remote areas, far from specialized centers [2]. The diversity of the different subtypes in RLCs and the proliferation of recombinant forms favored by the emergence of HIV resistance to treatment, which requires the choice of a technique capable of detecting several subtypes of HIV-1, sensitive, reproducible and less expensive. It is for this reason that in this study we will evaluate the sensibility of different techniques will be evaluated according to different subtypes of HIV-1 to recommend one of them for RLCs. Measurement of VL is used to assess the effectiveness of the treatment and to see the course of the disease. However, in order for the measurement to be significant, the difference between two results must be less than 0.5 log10, a difference from simple to triple. Thus, a VL of 100,000 RNA copies/ml (5 log10) and a VL of 250,000 RNA copies/ml (5.4 log10) are not considered to be significantly different [3,4]. Table 1 summarizes the 3 techniques used in this study [5,6]. Therefore, the aim of the study was to evaluate the sensitivity of different VL measurement techniques for HIV-1 non-B subtypes in N'Djamena in Chad.
| Assays | CAP/CTM V2.0 | Abbott | Generic HIV Viral Load® |
|---|---|---|---|
| Equipment for amplification/ detection | Cobas Taqman | m2000rt | Open platform or FluoroCycler |
| Principle | RT-PCR Real-Time | RT-PCR Real-Time | RT-PCR Real-Time |
| Kits | AmpliPrep/CobasTaqMan V2.0 | Abbott Real-time HIV-1 quantitative Assay: m2000system | PCR Real-Time in house |
| Amplified regions | gag and LTR | intégrase, gene and pol | LTR |
| Types and subtypes detected | M (A à G) + O | M (A à H) + O + N | M (A à H) |
| Quantitative threshold for detection (RNA copies/ml) | 20 | 40 | 390 |
| Number of tests/day (8 h per day) | 144 | 96-144 | 192 |
| Sample type | plasma | Plasma and DBS | Plasma and DBS |
Table 1: Comparative table of plasmatic viral load from the 3 techniques
Methods
One hundred and sixteen patients under ARV treatment, 78 women and 38 men representing respectively 67.24% and 32.76% recruited on the basis of specific inclusion criteria were enrolled in the study. The average age was 41 ± 6.87 years. Each patient signed an informed consent form after receiving sufficient information about the benefit of the study. The sampling used was consecutive and not exhaustive. The patients were recruited for 5 months (June-October 2013). A venous blood sample was taken at the elbow crease in two EDTA tubes of 5 ml each. The two tubes were stirred gently to mix the blood with the anticoagulant. The blood was centrifuged for 10 min at 2000 g. The plasma was aliquoted in three 2 ml cryotubes (2 to 3 cryotubes were used depending on the plasma volume). They are then put in a box and stored at -80°C and transported to the AIDS laboratory of Liège in Belgium for the analyzes. Three techniques were used to measure the VL on the collected samples of HIV-1 non-B subtypes infected patients. For CAP/CTM, plasma did not undergo freezing or thawing. But for the Abbott Real time and the Generic HIV Viral Load® assay, the plasma was subjected respectively one and two freezing/thawing.
Cobas ampliprep/cobas taqMan V2.0 (CAP/CTM)
COBAS® AmpliPrep/COBASTaqman® HIV-1 version 2.0 (CAP/CTM v2.0) is a test based on the in vitro amplification of HIV-1 RNA from plasma collected on EDTA tube. Introduced in 2009 [7,8] it seems to give better results than older versions, notably v1.0 and Amplicor v1.5 [9-11]. It can quantify 20 to 10,000,000 RNA copies/ml of HIV-1 M and O group. The reaction mixture designed to allow equivalent quantitative determination of HIV-1 M and O subtypes contains primers and Probes specific to both viral RNA and standard quantification RNA (QS), which uses reverse transcription and PCR amplification primers that define sequences in the highly conserved regions of the gag gene and The LTR region.
Extraction: A volume of 1000 μl of plasma is used for the extraction of the RNA while the machine draws only 850 μl. CAP/CTM v2.0 uses automated sample preparation and extraction on the COBAS AmpliPrep instrument using a generic silica-based capture technique.
Reverse transcription and PCR amplification: Reverse transcription and amplification are performed with the DNA polymerase of thermostable recombinant enzyme Thermus species (Z05). The reaction mixture is heated to allow the anti-sense primers to hybridize specifically to the HIV-1 target RNA and the HIV-1 QS RNA. In the presence of Mg2+ and an excess of dNTPs, the polymerase Z05 lengthens the hybridized primers, thus producing strands of DNA complementary to the target RNA.
Abbott real time test ref 2G3190: The Abbott Real Time HIV-1 assay in vitro is an RT-Real Time (Reverse Transcription in Real Time). A RT-qCR is a conventional PCR after reverse transcription of the RNA into cDNA. In other words, a PCR carried out on a cDNA obtained from an RNA by the action of a Reverse Transcriptase (RTase). Test on plasma and on whole blood collected on blotting paper on adult and pediatric samples. This test allows the detection of subtypes HIV-1: group M (A-H), groups N and O. Real-time PCR is based on the amplification and detection of a fluorescent reporter. The amount of HIV-1 target sequences present at each amplification cycle is measured using fluorescence-labeled oligonucleotide probes on the Abbott m2000rt apparatus. The amplification cycle in which the fluorescent signal is detected by the m2000rt is proportional to the log of the HIV-1 RNA concentration present in the original sample. The detection threshold is 40 RNA copies/ml.
Generic HIV viral load® assay (Biocentric, Bandol-France): It is of interest in the detection and quantification of HIV-1 RNA targeting the LTR gene, which is described as the least-changing region of the HIV-1 genome [12-14]. Biocentric markets the kit for the measurement of the VL described by Rouet and collaborators in the ANRS (Agence Nationale de Recherche sur le SIDA) HIV quantification working group. This Generic HIV Viral Load® assay allows the detection of most HIV-1 subtypes in group M [15]. This assay uses, unlike the other assays, extracted RNA for PCR amplification. The extraction kit used is the QIAamp DNA mini kit (Qiagen, Courtaboeuf, France). Hundred and Forty microliter of the plasma preserved at -80°C is used for extraction to obtain an eluate of 60μl at the end. Ten microliters of control (RNA virus) is placed in all the samples before extraction. For RT-PCR, a total volume of 25 μl containing 20 μl of Master Mix and 5 μl of RNA extract is used. The primers (forward and reverse: final concentration of 500 nM each), the probe (concentration of 200 nM) and Taqman One-Step RT-PCR Master Mix (Applied Biosystems). The primer used is the HIV1MGF 5'-GCCTCAATAAAGCTTGCCTTGA-3'. The sequence of the antisense primer is HIV1MGR 5'- GGCGCCACTGCTAGAGATTTT-3'. The probe used is HIV1MGProbe 5'-AAGTAGTGTGTGCCCGTCTGTTRTKTGACT-3'. The reporter for the probe is 5': 6-carboxyfluorescein and 3' quencher: 6-carboxytetramethylrhodamine (Applied Biosystems, Foster City, CA). The probe is labeled 5' with 6-FAM (Fluorochrome) and 3' with TAMRA (Quencher). The enzyme used is TaqMan one step. The standard curve is made by dilutions of 10 on one control, the Cy5 RNA Detection Probe. The detection threshold is 390 RNA copies/ml for 250 μL of plasma. For each assay, negative and positive control samples were used for all techniques.
Results
Comparison cobas ampliprep/cobas TaqMan V2.0 and abbot real time (n=110)
The VL was measured on all 116 samples first on CAP/CTM v2.0 and then on Abbott Real Time. The comparison was made on 110 samples tested successfully in both automates. Here, the plasma has not undergone freezing or thawing. The results of the VL measurement are given in the following Table 2.
| Viral Load | Cobas AmpliPrep/TaqMan V2.0 | Abbott RealTime | ||
|---|---|---|---|---|
| Copies/ml | Log10 | Copies/ml | Log10 | |
| Minimum | 0,00 | 0,00 | 0,00 | 0,00 |
| Maximum | 966 000 | 5,98 | 885 196 | 5,95 |
| Mean | 43 301 | 4,64 | 43 460 | 4,6 |
| Mediane | 85 | 1,93 | 51 | 1,7 |
| ≥ 1000 | 48 | 48 | ||
| < 1000 | 31 | 29 | ||
| Not detected | 32 | 37 | ||
| Invalid | 5 | 2 | ||
Table 2: Viral load measurement for 3 techniques (n=116)
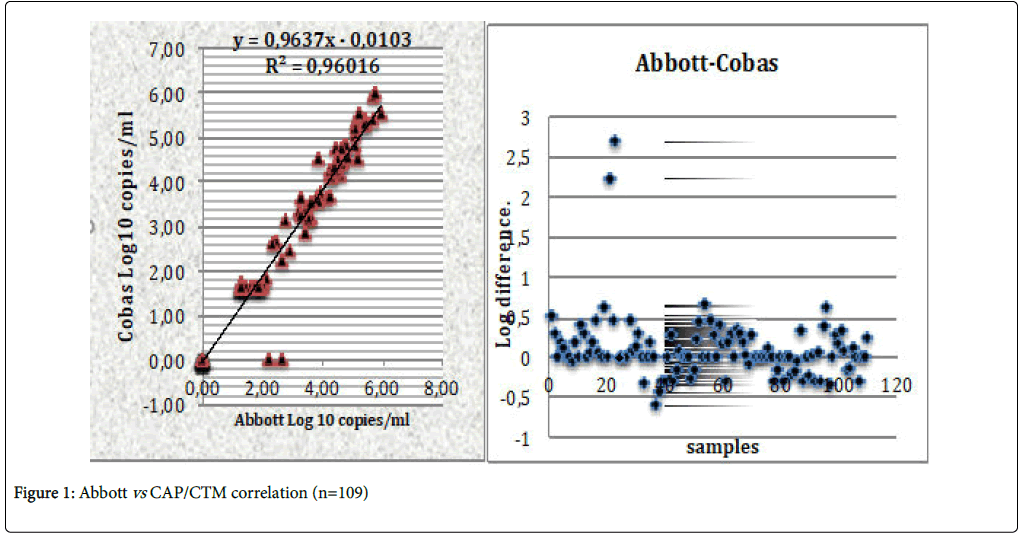
Invalid samples are due to the fact that the volume of the plasma is very insufficient, and the PLCs cannot read. Some have been supplemented with negative serum. It is important to remember that for two measurements of VL to be significant, the difference must be at least 0.5 log10. Figure 1 present the comparison of CAP/CTM v2.0 and Abbott Real Time. The distribution of points for Abbott and Cobas gives a satisfactory Pearson correlation coefficient (R2=0.96016). Therefore, these Abbott and Cobas measures are statistically similar (p<0.05) for α=5%. We noticed that two samples were detected with Abbott but were not detected with CAP/CTM v2.0. This would be due to a blip during sampling.
Comparison cobas ampliprep/cobas taqMan v2.0, abbott real time and generic HIV viral load® assay (n = 42)
For measurement with the Generic HIV Viral Load® assay, 42 samples were tested and compared with the Abbott and Cobas techniques (Table 3).
| Viral Load | Cobas | Abbott | Generic HIV Viral Load® | |||
|---|---|---|---|---|---|---|
| Copies/ml | Log10 | Copies/ml | Log10 | Copies/ml | Log10 | |
| Minimum | 20,00 | 1,3 | 40,00 | 1,6 | 390,00 | 2,6 |
| Maximum | 966000 | 5,98 | 885196 | 5,95 | 281453,19 | 5,56 |
| Mean | 104577,17 | 5,02 | 101963,22 | 5,01 | 24937,91 | 4,40 |
| Mediane | 30850 | 4,49 | 25762 | 4,41 | 5747,55 | 3,76 |
Table 3: Viral load measurement for 3 techniques (n=42)
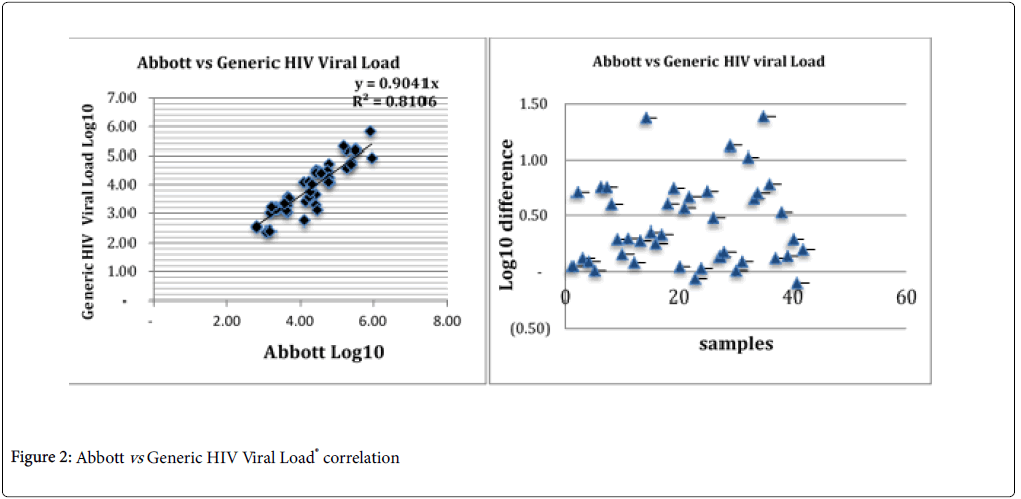
This table gives information on the different values measured by the three different techniques. We noted that there is no difference in the values between the minimum and maximum values (p<0.05). While for median and mean values, the Generic HIV Viral Load® measurement is different from at least 0.5 Log10. Therefore, it is significantly different from the first two (p>0.05). This would certainly be due to the fact that the plasma was thawed at least twice. The correlation between Generic HIV Viral Load® and Abbott is good with a Person correlation coefficient of R2 of 0.81064 (p<0.05) (Figure 2).
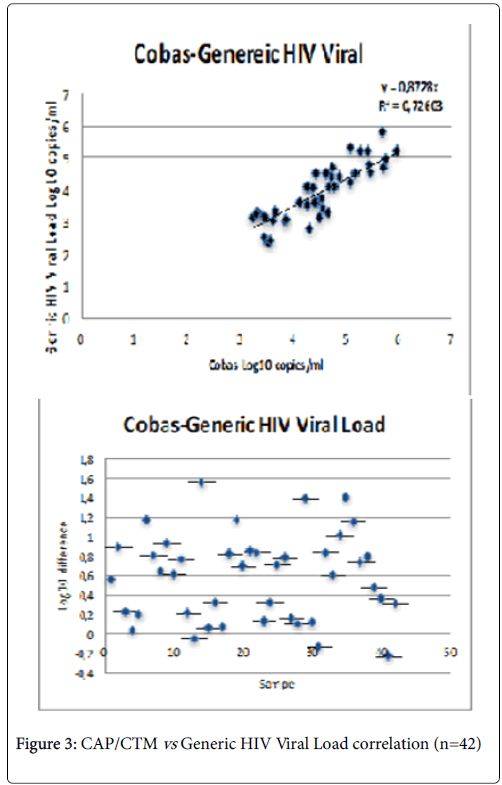
Good correlation between Cobas and Generic HIV Viral Load® with a Pearson R2 correlation coefficient of 0.72603 (p<0.05) (Figure 3). Given that each test used at a specific sensitivity to the different HIV-1 groups and subtypes, it goes without saying that some subtypes can be detected by some tests while others cannot.
Discussion
In Resources-Limited Countries (RLCs), the assessment of virological failure is often late according to the WHO recommendations [16,17] this is sometimes the cause of accumulation of resistance mutations and reduced effectiveness of second-line drugs. Notwithstanding, some authors report an urgent need for virological monitoring by measuring VL [18-20]. For others, the monitoring of HIV-1 VL in RLCs faces multiple challenges [7,21]. The ANRS assay marketed by Biocentric under the name “Generic HIV Viral load assay” is used routinely in Ivory Coast, Cambodia, Vietnam, Gabon, Cameroon. Various concordance evaluations for this technique with the Cobas Monitor V1.5 were done by Rouet et al. in various countries with an average correlation coefficient of 0.79 [22]. A study published in 2015 presented a correlation coefficient of 0.9452 between Generic HIV Viral Load® assay and the CAP/CTM v2.0 using 39 samples of non-B subtypes of HIV-1 [23]. In this study, comparing the Generic HIV Viral Load® assay with CAP/CTM v2.0, there was a correlation coefficient of 0.72603 with CAP/CTM v2.0 despite the fact that the plasma had undergone two thawing measure with the Generic HIV Viral Load® assay. The different values of this coefficient per country described by Rouet et al. for Generic HIV Viral Load® were: France (0.8701, n=88), Morocco (0.8624, n=50), Zimbabwe (0.8703, n=52), Cambodia and Thailand (0.8446, n=34), Central African Republic (0.8924, n=25) and Madagascar (0.6814; n = 22) [14]. Comparing the CAP/CTM v2.0 assay with Abbott Real-time PCR in South Africa, Lesley et al. obtained a good correlation coefficient R2=0.908 (p<0.05). As for Karasi et al. comparing the two Abbott m2000 Real-Time and CAP/CTM V2.0 assays for the B and non-B subtypes, obtained a correlation coefficient of R2=0.95 [24,25]. Wirden et al. compared Abbott and CAP/CTM on non-B subtypes and did not obtain a significant difference between the two with R2=0.84 [25]. Van Rensburg et al. in two cohorts made in Africa and the USA did not find significant differences between the two techniques with a strong correlation of 95% [26]. All these results are in the same direction as this study on these Abbott and Cobas methods because the correlation coefficient obtained in our study (R2=0.96016) corroborates well with those of these authors for Abbott and CAP/CTM v2.0. In a study by Margariti et al. in 2016, comparing low VL (<200 RNA copies/ml) in patients infected with subtypes B and non-B, noted a significant difference and concluded that for virological monitoring, a single technique between the two should be used [27]. In this study, for the measurement between CAP/CTM v2.0 and Abbott Real Time, two samples not detected in Abbott but detected in Cobas with 2.23 and 2.68 Log10 respectively. This variation could be due to a blip on two samples. A "blip" is defined as a transient elevation of plasma HIV RNA, usually between 50 and 1000 RNA copies/ml, observed on a single sample, and does not justify the prescription of a Resistance test [22]. Studies have shown that CAP/CTM v2.0 is an accurate and reliable measure of HIV-1 VL versus other assays used for genotype specificity and susceptibility [11].
As for the specificity and sensitivity of the techniques, we note that the CAP/CTM v2.0 values are better than those observed for Abbott Real Time. It must also be said that the sensitivity of the techniques differs from one subtype to another. Therefore, CAP/CTM v2.0 is only sensitive to subtypes A-H of group M. While Abbott Real Time detects the N, O groups and the A-G subtypes of the M group. Finally, the Generic HIV Viral Load® assay detects the sub-types from A to H of the M group [28]. The Pearson coefficients obtained with CAP/CTM v2.0 and Generic HIV Viral Load® assay (0.72603) are due to the freezing and thawing of the plasma more than once; this confirms the fact that plasma freezing, and thawing has more than twice interferes with the number of copies of the VL. The objective of the evaluation the performance of these different techniques of VL is precisely to have a great sensitivity and possibility of detection of the different subtypes in the countries with limited resources and notably in Chad.
Conclusion
The correlation between COBAS TaqMan/AmpliPrep and Abbott Real Time Viral Load is very good with R2 equal to 0.96016; while the measurement with the Generic HIV Viral Load® assay of the Generic HIV Viral Load® seems to be discordant of the first two but the difference of Log10 is not very significant. This discrepancy could be explained by the fact that for the measurement with the Generic HIV Viral Load® assay, the plasma was frozen and thawed twice. Abbott Real time remains the recommended technique for resource-poor countries, particularly Chad, because of its sensitivity and variability in detecting different subtypes of HIV-1.
Acknowledgments
The authors would like to thank the technicians of the laboratory of Virology of the General Hospital of Reference N’Djamena and those of the AIDS Reference Laboratory of the CHU of Liège for facilitating the data collection.
References
- Palella FJ, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, et al. (1998) Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N Engl J Med 338: 853-860.
- Adawaye C, Kamangu E, Ali Mahamat M, Tchoumbou HZB, Vaira D, et al. (2013) Use of dried blood spot to improve the diagnosis and management of HIV in resource-limited settings. World J AIDS 3: 251-256.
- Chirouze C, Hoen B (2006) Surveillance du patient sous traitement antir??troviral. Rev du Prat 56: 966-975.
- Greder Belan A, Chaplain C, Boussairi A (2008) Suivi biologique de l’infection?VIH chez l’adulte. Immuno-Analyse Biol Spec 23: 95-102.
- Gueudin M, Simon F (2005) Plasma RNA viral load in HIV-1 group O infection by real-time PCR. Methods Mol Biol 304: 221-228.
- Pilcher CD, Eron JJ, Vemazza PL, Battegay M, Harr T, et al. (2001) Sexual transmission during the incubation period of primary HIV infection. J Am Med Assoc 286: 1713-1714.
- Rouet F, Chaix ML, Nerrienet E, Ngo-Giang-Huong N, Plantier JC, et al. (2007) Impact of HIV-1 genetic diversity on plasma HIV-1 RNA quantification. J Acquir Immune Defic Syndr 45: 380-388.
- De Bel A, Marissens D, Debaisieux L, Liesnard C, Van Den Wijngaert S, et al. (2010) Correction of underquantification of human immunodeficiency virus type 1 load with the second version of the Roche Cobas AmpliPrep/Cobas TaqMan assay. J Clin Microbiol 48: 1337-1342
- Oliver AR, Pereira SF, Clark DA (2007) Comparative evaluation of the automated Roche TaqMan real-time quantitative human immunodeficiency virus type 1 RNA PCR assay and the Roche AMPLICOR version 1.5 conventional PCR assay. J Clin Microbiol 45: 3616-3619.
- Paba P, Fabeni L, Ciccozzi M, Perno CF, Ciotti M (2011) Performance evaluation of the COBAS/TaqMan HIV-1 v2.0 in HIV-1 positive patients with low viral load: A comparative study. J Virol Methods 173: 399-402.
- Ribas SG, Heyndrickx L, Ondoa P, Fransen K (2006) Performance evaluation of the two protease sequencing primers of the Trugene HIV-1 genotyping kit. J Virol Methods 135: 137-142.
- Drosten C, Panning M, Drexler JF, Hänsel F, Pedroso C, et al. (2006) Ultrasensitive monitoring of HIV-1 viral load by a low-cost real-time reverse transcription-PCR assay with internal control for the 5??? long terminal repeat domain. Clin Chem 52: 1258-1266.
- Rouet F, Rouzioux C (2007) The measurement of HIV-1 viral load in resource-limited settings: How and where? Clin Lab 53: 135-148.
- Rouet F, Ekouevi DK, Chaix ML, Burgard M, Inwoley A, et al. (2005) Transfer and evaluation of an automated, low-cost real-time reverse transcription-PCR test for diagnosis and monitoring of human immunodeficiency virus Type 1 infection in a west african resource-limited setting. J Clin Microbiol 43: 2709-2717.
- Rouet F, Foulongne V, Viljoen J, Steegen K, Becquart P, et al. (2010) Comparison of the Generic HIV Viral Load® assay with the AmplicorTM HIV-1 Monitor v1.5 and Nuclisens HIV-1 EasyQ® v1.2 techniques for plasma HIV-1 RNA quantitation of non-B subtypes: The Kesho Bora preparatory study. Journal of Virological Methods 163: 253-257.
- Mee P, Fielding KL, Charalambous S, Churchyard GJ, Grant AD (2008) Evaluation of the WHO criteria for antiretroviral treatment failure among adults in South Africa. AIDS 22: 1971-1977.
- Van Oosterhout JJG, Brown L, Weigel R, Kumwenda JJ, Mzinganjira D, et al. (2009) Diagnosis of antiretroviral therapy failure in Malawi: Poor performance of clinical and immunological WHO criteria. Trop Med Int Heal 14: 856-861.
- Calmy A, Ford N, Hirschel B, Reynolds SJ, Lynen L (2007) HIV viral load monitoring in resource-limited regions: Optional or necessary? Clin Infect Dis 44: 128-134.
- Harries AD, Zachariah R, van Oosterhout JJ, Reid SD, Hosseinipour MC, et al. (2010) Diagnosis and management of antiretroviral-therapy failure in resource-limited settings in sub-Saharan Africa: Challenges and perspectives. Lancet Infect Dis: 10: 60-65.
- Usdin M, Guillerm M, Calmy A (2010) Patient needs and pointâ€ofâ€care requirements for HIV load testing in resourceâ€limited settings. J Infect Dis 201: S73-S77.
- Fiscus SA, Cheng B, Crowe SM, Demeter L, Jennings C, et al. (2006) HIV-1 viral load assays for resource-limited settings. PLoS Med 3: 1743–1750.
- Wojewoda CM, Spahlinger T, Harmon ML, Schnellinger B, Li Q, et al. (2013) Comparison of Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 test version 2.0 (CAP/CTM v2.0) with other real-time PCR assays in HIV-1 monitoring and follow-up of low-level viral loads. J Virol Methods 187: 1-5.
- Kamangu EN, Chatte A, Boreux R, Susin F, Kalala RL, et al. (2015) Comparison of an in-house quantitative real-time PCR and COBAS ampliprep/taqMan roche for determination of viral load for HIV Type 1 Non-B. OALib 2: 1-7.
- Karasi JC, Dziezuk F, Quennery L, Förster S, Reischl U, et al. (2011) High correlation between the Roche COBAS® AmpliPrep/COBAS® TaqMan® HIV-1, v2.0 and the Abbott m2000 RealTime HIV-1 assays for quantification of viral load in HIV-1 B and non-B subtypes. J Clin Virol 52: 181-186.
- Wirden M, Tubiana R, Marguet F, Leroy I, Simon A, et al. (2009) Impact of discrepancies between the Abbott RealTime and Cobas TaqMan assays for quantification of human immunodeficiency virus type 1 group M non-B subtypes. J Clin Microbiol 47: 1543-1545.
- Van Rensburg EJ, Tait K, Watt A, Schall R (2011) Comparative evaluation of the roche cobas AmpliPrep/Cobas TaqMan HIV-1 version 2 test using the TaqMan 48 analyzer and the Abbott RealTime HIV-1 assay. J Clin Microbiol 49: 377-379.
- Margariti A, Chatzidimitriou D, Metallidis S, Pilalas D, Kourelis A, et al. (2006) Comparing Abbott m2000 RealTime HIV test and Roche COBAS Ampliprep/COBAS Taqman HIV test, v2.0 in treated HIV-1 B and non-B subjects with low viraemia. J Med Virol 88: 724-727.
- Hocini H, Andreoletti L (2009) Méthodes d’analyse et de suivi de l’infection par les virus de l’immunodéficience humaine. Rev Francoph des Lab: 39-48.
Citation: Adawaye C, Kamangu EK, Fokam J, Susin F, Moussa MA, et al. (2018) Comparison of 3 Tests for Plasma HIV-1 RNA Quantitation of Non-B Subtypes in Patients Infected with HIV-1 in N’Djamena-Chad: Cobas AmpliPrep/Cobas TaqMan HIV-1 Test Version 2.0, Abbott m2000 RealTime and Generic HIV Viral Load® Assays. J Infect Dis Ther 6: 382. DOI: 10.4172/2332-0877.1000382
Copyright: © 2018 Adawaye C, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 5534
- [From(publication date): 0-2018 - Nov 14, 2025]
- Breakdown by view type
- HTML page views: 4523
- PDF downloads: 1011