Research Article Open Access
Combining Ability Analysis for Northern Leaf Blight Disease Resistance on Tanzania Adapted Inbred Maize Lines
Tulole Lugendo Bucheyeki*1,2, Pangirayi Tongoona2, John Derera2 and Suzan Nchimbi-Msolla3
1Agricultural Research Institute (ARI)-Uyole, Mbeya, Tanzania
2African Crops Centre Institute (ACCI), University of KwaZulu-Natal, KwaZulu-Natal, South Africa
3Sokoine University of Agriculture (SUA), Morogoro, Tanzania
- *Corresponding Author:
- Tulole Lugendo Bucheyeki
Agricultural Research Institute (ARI)-Uyole
PO Box 400, Mbeya, Tanzania
Tel: +255 782237383
E-mail: tlbucheyeki@yahoo.co.uk
Received Date: March 09, 2017; Accepted Date: March 24, 2017; Published Date: March 31, 2017
Citation: Bucheyeki TL, Tongoona P, Derera J, Nchimbi-Msolla S (2017) Combining Ability Analysis for Northern Leaf Blight Disease Resistance on Tanzania Adapted Inbred Maize Lines. Adv Crop Sci Tech 5:266. doi: 10.4172/2329-8863.1000266
Copyright: © 2017 Bucheyeki TL, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Advances in Crop Science and Technology
Abstract
Northern leaf blight (NLB) disease incited by Exserohilum turcicum has increased in incidence and severity. Inbred lines combining ability and its interactions to the environment are required for the development of NLB disease resistance. The specific objectives were to estimate the combining ability for NLB disease resistance, determine maternal effects, and determine the heterosis in the F1 hybrids by using a full 11 × 11 diallel cross. The resulting 110 F1 hybrids with the 11 parents were evaluated together with 9 commercial varieties at three Agricultural Research Institutes: Tumbi, Uyole and Selian which represent diverse environments. Breeding materials were planted in 13 × 10 alpha lattice design with two replications per site. Top ten experimental hybrids in each site had negative mid parent heterosis for NLB disease severity. Heterosis for NLB disease severity ranged 94-362%. The overall mid parent heterosis means for yield across sites was 152%. Maternal effects had non-significant (P>0.005) influence on the inheritance of the NLB disease severity. Mean sum of squares for GCA was highly significant (P<0.001) on disease severity indicating additive gene action. Mean sum of squares for SCA were highly significant on disease severity and yield implying non-additive gene action. The mean squares for reciprocals effects were highly significant for yield and non-maternal effects sums of squares had significant effect (P<0.05) on yield. The GCA effects contribution was high for disease severity (91%) and lesion number (85%). With the exception of CML 395 and KS03-0B15-12 parents which were susceptible, all GCA effects were negative implying the contribution to disease resistance in their progenies. Due to preponderance of the additive gene action, recurrent selection could be used to improve the resistance of inbreeds while the non-additive gene action could be exploited in breeding for disease resistant high yielding hybrids.
Keywords
Combining ability; Maize; Northern leaf blight; Resistance; Tanzania
Introduction
Northern leaf blight disease (NLB) also known as Turcicum leaf blight or Northern corn leaf blight incited by the fungus Exserohilum turcicum (syn. Helminthosporium turcicum), Teliomorph Setophaeria turcica, is an endemic foliar maize disease in the world. The past decades witnessed the disease concentrating in the highlands of the world. However, now it affects both highlands and mid altitude maize growing areas and causing a significant yield reduction [1,2]. Maize yield losses caused by NLB disease vary depending upon location, pathogen virulence, pathogenicity, plant growth stage, number and position of leaves affected, relative humidity, temperature and susceptibility or resistance of the host plant. Raymundo [3] narrates the grain yield loss of over 50% in USA, while in India, the yield reductions of about 60% have been reported [4]. In East Africa, a maize grain yield reduction with the range of 16-24% have been reported [5].
Breeding for host plant resistance remains the most reliable and steadfast method of controlling the disease. Hence, host plant resistant (HPR) is considered as the best option and alternative to deal with the NLB disease problem [6,7]. However, breeding for HPR largely depends on correct methods of selecting suitable parents to be candidates of breeding for disease resistance. To study the potential, suitability and applicability of breeding materials, breeders apply different mating designs to achieve this purpose. One of the common mating designs is the diallel cross which have been extensively used in evaluations of breeding materials potential in various crops [8-12]. The diallel cross design enables breeders to estimate general combining ability (GCA) and specific combining ability (SCA) which are frequently used in genetic studies. Literature survey shows that, GCA for NLB disease is normally significant to denote its importance in additive gene action contribution. Vivek et al. [13] further contend that, GCA in NLB disease is area specific and is affected by environmental effects. This means that resistance to NLB disease varies from one location to another unless is monogenic before breaking down [13,14].
The majority of resistance of maize diseases is quantitatively inherited. In maize, NLB disease shows vertical and horizontal resistance inheritance mechanisms [15]. NLB disease is reported to be controlled by six dominant Ht1, Ht2, Ht3, HtN, NN and HtM and one recessive ht4 genes [16-20]. All these provide qualitative inheritance in the form of dominance or partial dominance. According to Pataky et al. [21] the HtN gene confers partial resistance to NLB disease. Other researchers have reported on the durable resistance to NLB conferred by major genes [22]. Ogliari et al. [23] reports on dominant HtP genes inducing resistance to NLB pathogen and recessive rt genes inducing resistance to specific NLB pathogen races. Several types of gene action are involved in controlling the inheritance of NLB disease in maize. Additive, dominance, and epistatic gene action have been reported in controlling the disease expression. However, additive gene action was found as the most important [23,24]. Maternal effects are less important for the traits associated with the inheritance of NLB. For example, Sigulas et al. [25] found non-significant maternal effect on 16 maize genotypes. Other researchers have reported non-significant importance of cytoplasmic and maternal effects on the inheritance of NLB resistance in maize genotypes [26].
Cultivars grown by farmers in Tanzania are potentially vulnerable to the NLB disease. Breeding for additional resistant varieties is needed. But, genetic information like GCA and SCA on the available inbred lines which is adapted to Tanzania conditions to be used as sources of NLB disease resistance and hybrid development is not known. Although various studies have been conducted in the world to estimate gene action related to NLB disease resistance and revealed useful information, these findings are limited to specific crosses and in some cases area bounded [27]. There is still potential room to widen the resistance genetic base as reports of new occurrence, distribution and resurgence of NLB disease are escalating worldwide. Another challenge facing the NLB disease struggle is the presence of pathogen races. The common known races include 0, 1, 2, 3, 4, 12, 23 and 23N. To make the matter more complex, there is emergence of new races. These create a new dimension to fight war against the pandemic. The recent studies in Kenya by Muiru et al. [28] revealed 0, 1, 2, 3, N, 12, 13, 13N, 3N, 123, 23 and 23N races to connote the constantly and non-stopping breeding for NLB disease in the area. Therefore, the study on NLB disease resistance was initiated in Tanzania. The overall objective was to study the gene action of NLB disease resistance in inbred maize lines adapted to Tanzania conditions. The specific objectives were to- 1) estimate the combining abilities for NLB resistance of 11 maize inbred lines adapted to Tanzania condition, 2) determine maternal effects which are involved in NLB disease resistance in maize germplasm and, 3) determine the heterosis in the F1 hybrids.
Materials and Methods
Sources and characteristics of breeding materials
Germplasm used in this study were obtained from screening for NLB disease resistance of 70 breeding materials at ARI-Tumbi in the growing season 2008/2009. The screening study resulted to the selection of 11 parents with various reaction types to NLB disease which were selected for the study (Table 1). Cob sizes were assessed by the developed scale of 1-5 where 1=very small, 2=small, 3=average, 4=big, and 5=excellent while NLB disease reactions assessment was performed according to procedures developed by Reid [29].
| Name | Source | Grain colour | Cob size (1-5) | R×n |
|---|---|---|---|---|
| KS03-0B15-126 | SARI-Tanzania | White | 4 | R |
| EB04-0A01-304 | SARI-Tanzania (QPM line) | White | 4 | R |
| CML 159 | CIMMYT-Mexico | White | 5 | R |
| KS03-0B15-2 | SARI-Tanzania | White | 4 | R |
| KS03-0B15-45 | SARI-Tanzania | White | 4 | R |
| VL 05616 | CIMMYT-Zimbabwe (Vivek) | White | 5 | R |
| KS03-0B15-47 | SARI-Tanzania | White | 5 | R |
| CML 395 | CIMMYT-Zimbabwe | White | 5 | S |
| KS03-0B15-12 | SARI-Tanzania | White | 4 | S |
| CML 442 | CIMMYT-Zimbabwe | White | 4 | R |
| CKL 05007-B-B | CIMMYT-Kenya (KARI) | White | 4 | R |
Where, Trt=treatment, R × n=reaction, Num=treatment number, cob size 1=very small, and 5=bigger.
Table 1: Characteristics of 11 parents used in a diallel mating.
F1 hybrid development
Breeding materials used in this study were developed from an 11 × 11 full diallel cross mating design. Crosses were performed at Selian Agricultural Research Institute (SARI) in Arusha in Tanzania. These crosses resulted in 55 F1 and 55 reciprocal families.
Field evaluation
The resulting 110 F1 progeny and 9 commercial hybrids (Kilima- ST, Kito-ST, Lishe-H1, Lishe-K1, Selian-H208, Selian-H308, Stuka-1, TMV-1 and Vumilia-k1) were planted in 13 × 10 alpha lattice design in three locations with two replications per site. The breeding materials were evaluated in three sites namely ARI-Tumbi, Selian and Uyole in the 2010/2011 growing season. One F1 hybrid CML 159 × VL 05616 was doubled per replication to make 130 maize genotypes for evaluation. An experiment involving 11 inbred materials was set adjacent to the hybrid trial to avoid inter-plot competition and was planted on the same day to avoid biasness [30]. Inbred lines were planted in RCBD with two replications per site. Maize was planted at a spacing of 0.75 m × 0. 30 m, which gives a plant population density of 44,444 plants per hectare. Basal fertilizer was applied at a rate of 40 Kg P ha-1 and Murate of Potash was applied at the rate of 40 Kg K2O ha-1. Top dressing was done by using UREA (46%) to make a recommended rate of 100 Kg N ha-1. Disease field assessment was conducted at about three weeks after silking of each particular genotype.
Inoculation procedures
To ensure uniform disease infestation, artificial inoculation was conducted according to procedures described by Reid [29] as follows- A sample grinder machine [Laboratory mill model-4, Thomas-Wiley, Thomas scientific (TM), and USA] was used to grind infected leaves into powder form. Leaves were collected from Tabora, Arusha and Mbeya, where trials were conducted and then mixed. A bazooka (Sistrunk Inoculators, Starkville, MS 39759) was used to apply the powder in the whorl of the plants. One dose of powder from the bazooka application amounted to 0.1 g of leaves powder. Two applications, one at 6-8 leaf stage and the second at 11-12 leaf stage were conducted. Furthermore, two rows of a spreader local variety (Situka-1) which is highly susceptible to NLB disease was planted around the field and after every 10 rows of the test materials. The uses of spreaders have been used successfully in screening germplasm for other disease studies [19]. In addition to NLB disease resistance, maize genotypes were assessed for other agronomic characteristics. Other collected agronomic data included- Days to 50% anthesis, days to 50% silking, plant height, ear height, husk cover, kernel type, grain texture and yield. Grain yield was estimated using the following formula-
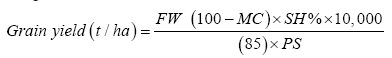
Where, FW=field weight of unshelled cobs (kg)
MC=grain moisture content (%)
SH%=Shelling percentage (expressed as a fraction)
10,000=1 ha=10,000 m2
PS=Plot area (m2)
85%=grain moisture content adjustment factor. Grain yield was adjusted to 15% (100-15). Data were recorded according to IBPGR [31] maize descriptors list. Yield and disease resistance data were validated and analyzed in Genstat statistical computer software [32].
Data analysis
Collected data were first analysed by analysis of variance [33] then by using Genstat computer software according to Payne et al. [34] to test the existence of significant differences on the measured maize traits. Gene action genetic components were estimated by application of SAS 05 computer programme using Griffing’s [9] Method 1, Model 1 (fixed model) as follows-

Yijk=value of F1 of a cross of the ith female and the jth male in the kth block and i plot/observation
μ=population mean; i=j=1, 2,…,n.
gi and gj=GCA effects of ith & jth parent
sij=SCA effect (sij=sji)
rij=reciprocal effects (rij=-rji)
eijkl=error effect for ijklth observation
b=number of replications
c=number of plants/plot
The ratio of GCA/SCA was also assessed. For the ratio greater than 1 indicated additive genetic effect while the ratio lowers than one denoted dominance gene action for the particular traits. The ratio closer to one implies the possibility of prediction based on GCA component only. The use of Griffing’s [9] Method 1, Model 1 provides similar results as other approaches [34,35].
Heterosis estimation
Mid parent heterosis was calculated according to Falconer and Mackay [36] and Saleh et al. [37] as follows:

Where, F1 =mean of the F1 hybrids; MPH=mid-parent heterosis and MP=mean of two parents.
Results
Estimation of general combining ability (GCA) and specific combining ability (SCA)
The model of analysis explained more than 60% of the variations (R2), R2 was 85, 64 and 73% for disease severity, yield and lesion numbers respectively to indicate that, the model was adequate (Table 2). The environment mean square was highly significant (P<0.001) for severity, yield and lesion number to imply the significant impact of environment on genotypes. The same trend was recorded on genotypes and on replications with the nested environment. The interaction of Environment and hybrids were highly (P<0.001) significant on disease severity only.
| Severity (%) | Yield | Lesion number | ||||
|---|---|---|---|---|---|---|
| Source | DF | MS | MS | DF | MS | |
| Env. | 2 | 8846.89*** | 92.37*** | 1241.12*** | ||
| Rep (Env.) | 3 | 1675.42*** | 86.82*** | 765.43*** | ||
| Genotypes | 120 | 770.3*** | 11.72*** | 74.28*** | ||
| Env. Genotypes | 240 | 110.26*** | 2.19 | 63.9 | ||
| Error | 360 | 68.89 | 3.75 | 68.89 | ||
| Corrected total | 725 | |||||
| R2 | 0.85 | 0.64 | 0.73 | |||
*, **, *** indicates significance level at 0.05, 0.01 and 0.001, respectively.
Table 2: Mean squares partial analysis of variance on NLB disease severity (%), yield and lesion number of maize hybrids.
Mean sum of squares of GCA was highly significant (P<0.001) for disease severity and lesion numbers and significant on yield (P<0.05) while SCA mean squares were highly significant for disease severity and yield only (Table 3). Highly significant maternal effects (P<0.001) was recorded on yield only. At the same time mean squares for reciprocals were highly significant for yield and non-maternal sum of squares had significant effect (P<0.05) on yield. In addition, % GCA contribution much higher for disease severity (90.81%) and lesion number (85.41%). However, % GCA contribution for yield was (8.71%) to indicate nonadditive gene action effects.
| Severity (%) | Yield | Lesion number | |||||
|---|---|---|---|---|---|---|---|
| Observation | Source | MS | MS | MS | |||
| 1 | GCA | 8078.13*** | 8.72* | 672.41*** | |||
| 2 | SCA | 148.67*** | 16.62*** | 20.89 | |||
| 3 | Reciprocal | 63.23 | 7.37*** | 18.93 | |||
| 4 | Maternal | 109.58 | 15.59*** | 40.22 | |||
| 5 | Non-maternal | 52.93 | 5.55* | 14.2 | |||
| 6 | GCA ×Env. | 553.23*** | 2.12 | 474.92*** | |||
| 7 | SCA ×Env. | 77.75 | 2.45 | 30.57 | |||
| 8 | Reciprocal ×Env. | 62.23 | 1.93 | 22.49 | |||
| 9 | Maternal ×Env. | 86.67 | 1.29 | 41.97 | |||
| 10 | Non-maternal ×Env. | 56.79 | 2.08 | 18.16 | |||
| %GCA contribution (ss) | 90.81 | 8.71 | 85.41 | ||||
| %SCA contribution (ss) | 9.19 | 91.29 | 14.59 | ||||
*, **, *** indicates significance level at 0.05, 0.01 and 0.001, respectively.
Table 3: Diallel analyses of hybrids for disease reaction and yield over three sites.
Table 4, shows the GCA effects of 11 parents used in NLB disease resistance study. All GCA effects had highly significant (P<0.001) effects on disease severity. With the exception of CML 395 and KS03-0B15-12, all CGA effects were negative. Parent EB04-0A01-304 had significant positive GCA effects on yield while VL 05616 and CKL 05007-B-B had significant negative GCA effects on yield (P<0.05). On lesion number, two parents, CML 395 and KS03-0B15-12 had highly significant positive GCA effects on lesion number (P<0.001), while parent CML 442 had negative significant (P<0.05) effects on the same trait.
| Parent | Severity (%) | GCA | Yield | GCA | Lesion number | GCA |
|---|---|---|---|---|---|---|
| KS03-0B15-126 | 9.92 | -3.66*** | 2.07 | 0.1 | 4.67 | -1.15 |
| EB04-0A01-304 | 12.03 | -3.74*** | 2.84 | 0.44** | 4.67 | -0.89 |
| CML 159 | 13.82 | -4.05*** | 2.46 | 0.24 | 5.33 | -1.12 |
| KS03-0B15-2 | 11.78 | -2.95** | 2.07 | -0.09 | 4.67 | -0.79 |
| KS03-0B15-45 | 15.88 | -3.27*** | 2.75 | -0.13 | 5.17 | -0.87 |
| VL 05616 | 11.6 | -2.40** | 2.48 | -0.35* | 5.83 | -0.6 |
| KS03-0B15-47 | 14.72 | -3.46*** | 2.29 | 0.25 | 4.33 | -1.43 |
| CML 395 | 29.2 | 14.61*** | 2.19 | 0.05 | 7 | 3.87*** |
| KS03-0B15-12 | 46.93 | 16.88*** | 3.39 | -0.22 | 6.5 | 5.14*** |
| CML 442 | 11.9 | -3.24*** | 2.37 | 0.04 | 5 | -1.27* |
| CKL 05007-B-B | 10.35 | -4.71*** | 2.56 | -0.36* | 3.83 | -0.87 |
*, **, *** indicates significance level at 0.05, 0.01 and 0.001 respectively.
Table 4: Combined general combining ability (GCA) effects for disease reaction and yield of parents over three sites.
In all three sites, All GCA had highly significant (P<0.001) effects on disease severity. With the exception to CML 395 and KS03-0B15-12, all CGA effects were negative. The same parents had significant positive GCA (P<0.001) effects on yield (Table 5).
| Tumbi | Uyole | Selian | Tumbi | Uyole | Selian | |
|---|---|---|---|---|---|---|
| Severity | Yield | |||||
| Parents | ||||||
| KS03-0B15-126 | -4.16*** | -4.40*** | -4.99*** | -4.41 | -1.59 | -1.18 |
| EB04-0A01-304 | -4.90*** | -4.77*** | -5.01*** | -2.29 | -1.44 | -0.93 |
| CML 159 | -4.31*** | -4.65*** | -4.56*** | -2.27 | -1.28 | -0.96 |
| KS03-0B15-2 | -4.19*** | -3.26*** | -3.17*** | -1.57 | -1.16 | -0.86 |
| KS03-0B15-45 | -4.15*** | -2.76*** | -2.67*** | -1.39 | -0.97 | -0.76 |
| VL 05616 | -3.02*** | -4.15*** | -4.06*** | -2.19 | -1.57 | -1.27 |
| KS03-0B15-47 | -5.12*** | -4.95*** | -4.85*** | -2.24 | -1.51 | -1.02 |
| CML 395 | 18.81*** | 19.26*** | 19.34*** | 9.71*** | 6.47*** | 4.88*** |
| KS03-0B15-12 | 20.59*** | 19.66*** | 19.76*** | 9.67*** | 6.53*** | 4.78*** |
| CML 442 | -4.14*** | -4.83*** | -4.74*** | -2.34 | -1.55 | -1.15 |
| CKL 05007-B-B | -4.17*** | -3.19*** | -3.01*** | -2.27 | -1.32 | -0.98 |
Table 5: Site combining ability effects for NLB disease severity and yield.
Three hybrids CML 159 × CML 395, VL 05616 × KS03-0B15-12 and KS03-0B15-2 × CML 395 had positive significant (P<0.05) SCA effects on disease severity while hybrid CML 395 × CKL 05007-B-B possessed positive highly significant SCA effects on yields (Table 6). Parents KS03-0B15-12 and CML 395 were susceptible to NLB disease. On yield, hybrid VL 05616 × KS03-0B15-12 had positive significant SCA effects (P<0.05) while hybrid CML 395 × CKL 05007-B-B expressed positive highly significant SCA (P<0.001) effects. Positive significant SCA is desired in yield of maize trait. All hybrids had non-significant SCA effects on lesion numbers.
| Cross | Severity (%) | Yield tha-1 | Height (cm) | Ear height (cm) | Anthesis days | Silking days | SCA (severity) | SCA (yield) | SCA (lesion) |
|---|---|---|---|---|---|---|---|---|---|
| 3 × 8 | 33.42 | 5.93 | 206.17 | 103.33 | 71.00 | 72.83 | 7.89** | 0.11 | 0.02 |
| 6 × 9 | 35.67 | 6.403 | 161.67 | 74.5 | 69.67 | 71.33 | 6.40* | 1.45** | 1.15 |
| 8 × 11 | 35.8 | 5.323 | 190 | 86.67 | 70.00 | 71.67 | 20.08*** | 3.39*** | 0.83 |
| 4 × 8 | 38.3 | 6.23 | 164.17 | 74.83 | 68.67 | 70.67 | 7.71** | 0.53 | 3.2 |
*, **, *** indicates significance level at 0.05, 0.01 and 0.001 respectively, Where, 3=CML 159, 4=KS03-0B15-2, 6=VL 05616, 8=CML 395 and 9=KS03-0B15-12,
Table 6: Means and specific combining ability (SCA) effects of crosses for NLB disease reaction and yield over three sites.
Reciprocal, maternal, nonmaternal effects on yield
Mean squares of SCA and maternal effects had highly significant positive (P<0.001) effects on yield while, GCA showed positive significant (P<0.005) effects on yield (Table 7). Reciprocal effects were also highly significant (P<0.001). The hybrids with positive highly significant effects were KS03-0B15-126 × KS03-0B15-12 and CML 159 × KS03-0B15-12 indicating the contribution of cytoplasmic gene and nuclear gene effects. Maternal effects were also observed. Five parents showed positive significant maternal effects at different dosage levels. For example parent KS03-0B15-126 showed positive highly significant (P<0.001) while KS03-0B15-47 showed positive significant (P<0.05) maternal effects. Nonmaternal positive significant effects were observed in four hybrids. With the exception of KS03-0B15-126 × KS03-0B15-12 which showed positive highly significant effects (P<0.001), the remaining EB04-0A01-304 × KS03-0B15-45, EB04-0A01-304 × KS03-0B15-45 and CML 159 × KS03-0B15-12 showed positive significant (P<0.05) maternal effects.
| Observation | Source | DF | MS |
|---|---|---|---|
| 1 | GCA | 10 | 8.72* |
| 2 | SCA | 55 | 16.62*** |
| 3 | Reciprocal | 55 | 7.37*** |
| 4 | Maternal | 10 | 15.59*** |
| 5 | Non-maternal | 45 | 5.54* |
| 6 | GCA × Environment | 20 | 2.12 |
| 7 | SCA × Environment | 110 | 2.45 |
| 8 | Reciprocal × Environment | 110 | 1.93 |
| 9 | Maternal × Environment | 20 | 1.29 |
| 10 | Non-maternal ×Environment | 90 | 2.07 |
| Cross/parent | Yield(tha-1) | Effects | |
| Reciprocal | 1 × 9 | 3.56 | -2.08*** |
| 3 × 6 | 5.17 | -1.22* | |
| 3 × 9 | 7.06 | 2.09*** | |
| 9 × 7 | 7.19 | 1.12* | |
| 5 × 8 | 2.88 | -1.32* | |
| 6 × 10 | 6.28 | 1.13* | |
| 8 × 10 | 7.61 | 1.61** | |
| Maternal | 1 | 2.07 | -0.55*** |
| 2 | 2.84 | 0.4* | |
| 7 | 2.29 | -0.32* | |
| 10 | 2.37 | -0.38* | |
| 11 | 2.56 | 0.4* | |
| Non- maternal | 1 × 9 | 3.56 | -1.7*** |
| 2 × 5 | 5.71 | -1.23* | |
| 3 × 6 | 5.17 | -1.3** | |
| 3 × 9 | 7.06 | 1.66* |
*, **, *** indicates significance level at 0.05, 0.01 and 0.001 respectively.
Table 7: Reciprocal, maternal, non-maternal effects on yield of parents and hybrids over three sites.
Heterosis
The studied F1 hybrids showed variations in the enhanced expression of mid parent heterosis (MPH) for NLB disease severity. At Tumbi, the mean was 12.97% and it was ranged 93.46-361.99%, while at Uyole, the mean was 13.22% and ranged 92.80-178.81%. At Selian, the mean was 11.14% and the ranged 76.60-144.19%. Each site ranked different F1 hybrids to indicate that, The F1 hybrids performed differently across sites (Table 8). The maximum mid parent heterosis for yield was higher than 350% in all sites.
| Tumbi | Uyole | Selian | |||
|---|---|---|---|---|---|
| Top ten | |||||
| 7×5 | -93.46 | 10×5 | -92.80 | 2 ×10 | -76.60 |
| 11×2 | -91.06 | 6×4 | -91.45 | 3 ×5 | -76.43 |
| 11×4 | -90.96 | 11×4 | -90.96 | 7 ×3 | -75.47 |
| 11×8 | -89.89 | 4×5 | -85.54 | 3 ×2 | -72.92 |
| 7×4 | -84.91 | 5×4 | -85.54 | 1 × 3 | -70.51 |
| 8×1 | -84.66 | 6×3 | -84.26 | 10×3 | -68.90 |
| 10×6 | -82.98 | 6×10 | -82.98 | 11×10 | -68.54 |
| 11×6 | -81.78 | 11×6 | -81.78 | 11×7 | -68.09 |
| 6×1 | -81.41 | 5×3 | -79.80 | 11 ×1 | -65.47 |
| 7×3 | -78.98 | 3×10 | -76.67 | 4 ×11 | -62.04 |
| Middle ten | |||||
| 5×6 | -41.78 | 4×10 | 1.35 | 5 ×10 | 7.99 |
| 4×10 | -40.88 | 2×6 | 1.57 | 5 ×1 | 8.53 |
| 2×1 | -36.22 | 9×11 | 4.75 | 2 ×1 | 9.34 |
| 11×7 | -36.18 | 11×9 | 4.75 | 2 ×7 | 9.91 |
| 9×10 | -35.41 | 9×8 | 5.08 | 9 ×1 | 10.82 |
| 10×9 | -35.41 | 1×9 | 5.54 | 5×9 | 11.45 |
| 3×9 | -34.16 | 7×6 | 6.38 | 6×1 | 11.52 |
| 10×7 | -32.38 | 10×11 | 7.87 | 8×7 | 11.57 |
| 7×9 | -31.87 | 5×10 | 7.99 | 8×2 | 11.57 |
| 11×1 | -30.93 | 3×6 | 10.15 | 7×9 | 13.54 |
| Bottom ten | |||||
| 8×7 | 95.81 | 10×6 | 104.26 | 11×8 | 87.10 |
| 10×8 | 118.98 | 1× 3 | 110.61 | 2×8 | 89.18 |
| 4×7 | 126.42 | 6×5 | 118.34 | 1× 4 | 93.55 |
| 6×8 | 145.10 | 1×11 | 126.94 | 4×8 | 104.98 |
| 3×11 | 148.24 | 1×8 | 130.06 | 8×4 | 104.98 |
| 5×7 | 161.44 | 3×8 | 132.45 | 3×8 | 111.53 |
| 4×5 | 189.23 | 8×3 | 132.45 | 8×11 | 112.39 |
| 1×11 | 196.00 | 8×6 | 145.10 | 5×8 | 121.83 |
| 7×10 | 200.53 | 8×11 | 152.84 | 10×8 | 128.71 |
| 2×4 | 361.99 | 1×6 | 178.81 | 1×5 | 144.19 |
| Statistics | |||||
| Mean | -12.97 | 13.22 | 11.14 | ||
| SD | 76.49 | 63.74 | 50.67 | ||
| Minimum | -93.46 | -92.80 | -76.60 | ||
| Ma×imum | 361.99 | 178.81 | 144.19 | ||
| Grand mean | 3.80 | ||||
Where, 1=KS03-0B15-126, 2=EB04-0A01-304, 3=CML 159, 4=KS03-0B15-2, 5=KS03-0B15-45, 6=VL 05616, 7=KS03-0B15-47, 8=CML 395, 9=KS03-0B15-12, 10=CML 442 and 11=CKL 05007-B-B
Table 8: Mid parent NLB disease severity heterosis across three sites.
At Tumbi, the mean was 133.60 and ranged 27.13-367.37 while at Uyole, the mean was 141.62 and ranged 45.30-352.98. At Selian, the mean was 180.32 and the ranged 12.55-460.84. The F1 hybrids behaved differently in terms of yield heterosis among sites (Table 9).
| Top ten | Tumbi | Uyole | Selian | ||
|---|---|---|---|---|---|
| 7×3 | 367.37 | 3×5 | 352.98 | 2×7 | 460.84 |
| 5×1 | 310.79 | 4×10 | 350.45 | 4×8 | 392.95 |
| 8×10 | 268.42 | 3×10 | 337.27 | 8×4 | 342.11 |
| 3×1 | 257.62 | 3×1 | 332.67 | 4×7 | 329.14 |
| 8×7 | 248.21 | 6×3 | 304.86 | 6×1 | 317.86 |
| 2×1 | 242.16 | 8×7 | 301.79 | 3×5 | 310.59 |
| 2×7 | 239.18 | 4×5 | 295.02 | 10×1 | 307.02 |
| 4×8 | 238.03 | 8×1 | 294.37 | 10×4 | 305.63 |
| 8×4 | 238.03 | 5×10 | 275.00 | 3×8 | 305.63 |
| 11×7 | 234.02 | 4×1 | 274.88 | 8×10 | 300.82 |
| Middle ten | |||||
| 10×8 | 150.00 | 9×7 | 153.52 | 1× 4 | 200.00 |
| 1×7 | 147.71 | 6×7 | 151.57 | 4×9 | 197.25 |
| 4×11 | 146.22 | 2×5 | 150.45 | 5×3 | 195.46 |
| 11×1 | 146.22 | 2×9 | 147.83 | 1×10 | 195.46 |
| 4×2 | 144.40 | 8×9 | 143.73 | 11×5 | 193.28 |
| 8×5 | 142.92 | 4×11 | 141.90 | 3×9 | 191.50 |
| 2×10 | 141.84 | 1×5 | 140.66 | 2×6 | 190.21 |
| 7×2 | 133.92 | 8×2 | 138.57 | 4×3 | 180.70 |
| 1× 4 | 131.88 | 7×1 | 138.53 | 1×8 | 178.26 |
| 4×9 | 130.77 | 7×4 | 138.53 | 7×9 | 176.92 |
| Bottom ten | |||||
| 5×11 | 24.29 | 8×11 | 9.47 | 10×5 | 49.15 |
| 9×3 | 23.08 | 7×11 | 7.22 | 6×5 | 47.69 |
| 6×4 | 18.68 | 9×1 | 6.96 | 4×6 | 42.42 |
| 10×9 | 14.58 | 9×4 | 2.56 | 10×11 | 37.50 |
| 11×3 | 7.57 | 9×2 | -3.69 | 11×10 | 29.08 |
| 9×6 | 2.21 | 3×11 | -20.32 | 9×5 | 22.66 |
| 7×11 | -1.03 | 6×4 | -20.88 | 9×3 | 18.76 |
| 5×10 | -6.25 | 5×11 | -32.20 | 6×4 | 12.50 |
| 3×11 | -16.33 | 5×8 | -35.22 | 10×9 | 0.40 |
| 5×8 | -27.13 | 9×3 | -45.30 | 9×6 | -12.55 |
| Statistics | |||||
| Mean | 133.60 | 141.62 | 180.32 | ||
| SD | 76.96 | 87.74 | 92.35 | ||
| Minimum | -27.13 | -45.30 | -12.55 | ||
| Ma×imum | 367.37 | 352.98 | 460.84 | ||
| Grand mean | 151.84 | ||||
Where, 1 = KS03-0B15-126, 2 = EB04-0A01-304, 3= CML 159, 4 = KS03-0B15-2, 5 = KS03-0B15-45, 6 = VL 05616, 7 = KS03-0B15-47, 8 = CML 395, 9 = KS03- 0B15-12, 10= CML 442 and 11 = CKL 05007-B-B
Table 9: Mid parent NLB disease yield heterosis across three sites.
Yield performance and NLB disease reactions of parent materials
There were no significant differences (P>0.05) in yield performance across test sites implying stability of inbred lines across environments (Table 10). On severity, testing environments differed significantly (P<0.001) with ARI Selian site recording the highest severity (22.9%) followed by ARI-Uyole 17.95%. Genotype performance across sites showed significant difference (P<0.001) on NLB disease lesion number with ARI-Uyole recording the highest (8.68) while ARI-Tumbi showed the lowest (1.55). On lesion length, there were also highly significant differences (P<0.001) across sites with ARI-Selian recording the highest (156.5 mm) followed by ARI-Uyole 26.1 mm).
| Yield tha-1 | Severity (%) | Lesion number | Lesion length(mm) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Tumbi | Uyole | Selian | Tumbi | Uyole | Selian | Tumbi | Uyole | Selian | Tumbi | Uyole | Selian |
| KS03-0B15-126 | 1.8 | 2.8 | 1.6 | 10.5 | 8.5 | 10.75 | 2.5 | 9 | 2.5 | 11 | 17.5 | 57.5 |
| EB04-0A01-304 | 3.6 | 1.31 | 3.6 | 0 | 13 | 23.1 | 0 | 10 | 4 | 0 | 27 | 100 |
| CML 159 | 2.55 | 2.28 | 2.55 | 14.5 | 10.5 | 16.45 | 1.5 | 9 | 5.5 | 8.5 | 22.5 | 143 |
| KS03-0B15-2 | 2.55 | 1.1 | 2.55 | 7.5 | 10 | 17.85 | 1 | 7.5 | 5.5 | 7.3 | 19.5 | 275.5 |
| KS03-0B15-45 | 2.62 | 3 | 2.62 | 5 | 18.5 | 24.15 | 1 | 10 | 4.5 | 2.5 | 40.5 | 159 |
| VL 05616 | 2.1 | 2.64 | 2.7 | 6 | 12 | 16.8 | 1.5 | 10 | 6 | 2.4 | 27 | 192 |
| KS03-0B15-47 | 2.1 | 2.68 | 2.1 | 16 | 14.5 | 13.65 | 1 | 8 | 4 | 2.5 | 27 | 60 |
| CML 395 | 2.55 | 2.42 | 1.6 | 15 | 32 | 40.6 | 4 | 10 | 7 | 12.8 | 40.5 | 135 |
| KS03-0B15-12 | 2.7 | 4.78 | 2.7 | 38 | 51 | 51.8 | 4 | 8.5 | 7 | 12 | 19.5 | 193 |
| CML 442 | 3.15 | 2.12 | 1.85 | 0 | 17.5 | 18.2 | 0 | 8.5 | 6.5 | 0 | 35.5 | 180 |
| CKL 05007-B-B | 3.3 | 1.44 | 2.95 | 2.5 | 10 | 18.55 | 0.5 | 5 | 6 | 3.5 | 10.5 | 226 |
| Mean | 2.64 | 2.42 | 2.44 | 10.45 | 17.95 | 22.9 | 1.55 | 8.68 | 5.32 | 5.7 | 26.1 | 156.5 |
| Lsd | 0.756 | 4.014 | 1.074 | 33.4 | ||||||||
| Cv | 49.3 | 38.2 | 33.8 | 86.7 | ||||||||
| Prob | 0.805 | <0.001 | <0.001 | <0.001 | ||||||||
Table 10: Yield and disease reaction of 11 inbred lines over three sites.
Yield performance of hybrids
The yield performance of experimental F1 hybrids and commercial hybrids did not differ (P>0.05) significantly across sites (Table 11). However on NLB disease severity, breeding materials showed highly significant (P<0.001) differences across sites. All commercial hybrids showed susceptibility to NLB disease with the scale of more than five. At ARI-Tumbi, the range was 4.85-6.18, while at Uyole the range was 5.90-7.19. At the same time the range at Selian was 5.08-6.93. Based on NLB resistance ranking performance of hybrids, there were no hybrids which performed consistently across sites (Table 12). However, on NLB disease resistance mean, ARI-Tumbi had the lowest (3.03) followed by Selian (4.38) and Uyole (4.39). For experimental hybrids, the majority showed resistance to NLB disease reactions. For example the range at Tumbi was 1.57-2.55, while at Uyole, the range was 1.37-7.24 and at Selian, the range was 1.83-6.97 of the square root transformed NLB disease severity percentage.
| Tumbi | Uyole | Selian | |||
|---|---|---|---|---|---|
| Top ten | Yield | Yield | Yield | ||
| 7×11 | 4.65 | 6×10 | 5.30 | 7×3 | 9.54 |
| 11×4 | 5.85 | 1×7 | 6.70 | 3×7 | 9.18 |
| 6×5 | 3.75 | 8×5 | 7.32 | 2×10 | 8.82 |
| 7×5 | 6.30 | 11×6 | 4.76 | 8×10 | 8.82 |
| 9×11 | 4.35 | 7×10 | 4.16 | 8×7 | 8.82 |
| 4×5 | 5.70 | 2×10 | 8.84 | 10×4 | 8.46 |
| 8×2 | 5.40 | 3×2 | 7.20 | 2×4 | 8.28 |
| 3×2 | 4.80 | 5×4 | 3.90 | 9×1 | 8.28 |
| 5×9 | 5.10 | 4×5 | 8.16 | 11×6 | 8.10 |
| 3×10 | 6.45 | 11×10 | 7.02 | 2×7 | 7.92 |
| Middle five | |||||
| 9×4 | 3.00 | 9×5 | 7.20 | 2×5 | 6.12 |
| 10×4 | 7.05 | 10×4 | 6.70 | 3×6 | 6.12 |
| 8×1 | 6.00 | 1×5 | 5.90 | 5×4 | 6.12 |
| 9×5 | 4.50 | 7×1 | 5.72 | 5×9 | 6.12 |
| 4×3 | 3.75 | 8×4 | 5.50 | 6×8 | 6.12 |
| Bottom five | |||||
| 5×10 | 3.90 | 8×11 | 3.76 | 9×3 | 3.60 |
| 3×7 | 7.65 | 10×1 | 3.70 | 9×4 | 3.60 |
| 7×6 | 4.20 | 7×8 | 7.40 | 1×9 | 3.43 |
| 4×8 | 6.30 | 11×4 | 6.80 | 3×11 | 3.24 |
| 4×6 | 4.50 | 6×8 | 5.84 | 5×8 | 2.70 |
| Commercial checks | |||||
| Kilima-ST | 6.90 | 5.40 | 5.16 | ||
| Kito-ST | 2.25 | 2.15 | 3.15 | ||
| Lishe-H1 | 3.75 | 6.80 | 5.25 | ||
| Lishe-K1 | 3.30 | 4.92 | 5.62 | ||
| Selian-H208 | 5.85 | 7.70 | 6.19 | ||
| Selian-H308 | 4.20 | 6.30 | 5.88 | ||
| Stuka-1 | 3.40 | 5.32 | 4.76 | ||
| TMV-1 | 4.20 | 5.32 | 5.81 | ||
| Vumilia-k1 | 5.70 | 5.90 | 5.48 | ||
| Statistics | |||||
| Mean | 5.04 | 6.31 | 6.05 | ||
| Lsd | 3.348 | 4.377 | 3.895ns | ||
| Cv | 33.60 | 35 | 32.5 | ||
| Prob | 0.542ns | 0.293ns | 0.581ns | ||
Where, 1=KS03-0B15-126, 2=EB04-0A01-304, 3=CML 159, 4=KS03-0B15-2, 5=KS03-0B15-45, 6=VL 05616, 7=KS03-0B15-47, 8=CML 395, 9=KS03-0B15-12, 10=CML 442 and 11=CKL 05007-B-B
Table 11: yield (tha-1) performance of hybrids across three sites.
| Tumbi | Uyole | Selian | |||
|---|---|---|---|---|---|
| Top ten | |||||
| 7×1 | 1.57 | 11×4 | 1.37 | 11×3 | 1.83 |
| 11×6 | 1.41 | 4×5 | 1.57 | 1× 3 | 2.05 |
| 5×4 | 2.09 | 10×5 | 1.62 | 4×11 | 2.14 |
| 11×7 | 2.41 | 5×7 | 1.71 | 11×1 | 2.16 |
| 10×2 | 2.85 | 6×10 | 1.83 | 3×10 | 2.28 |
| 3×7 | 1.62 | 11×7 | 1.98 | 11×7 | 2.32 |
| 3×1 | 1.5 | 3×7 | 1.98 | 11×10 | 2.5 |
| 1×7 | 1.5 | 4×11 | 2.09 | 11×4 | 2.51 |
| 8×7 | 5.34 | 4×2 | 2.12 | 7×3 | 2.52 |
| 1× 4 | 2.3 | 1×7 | 2.16 | 3×5 | 2.56 |
| Middle four | |||||
| 4×9 | 2.03 | 3×1 | 3.74 | 4×2 | 3.65 |
| 2×7 | 3.57 | 3×6 | 3.74 | 4×3 | 3.65 |
| 10×6 | 1.71 | 10×6 | 3.77 | 10×7 | 3.65 |
| 3×5 | 2.16 | 2×11 | 3.81 | 1×7 | 3.67 |
| Bottom four | |||||
| 10×1 | 1.98 | 8×9 | 6.98 | 8×11 | 6.84 |
| 5×3 | 1.87 | 8×6 | 7.07 | 5×8 | 6.86 |
| 10×7 | 2.21 | 6×9 | 7.1 | 3×8 | 6.87 |
| 2×1 | 2.55 | 8×3 | 7.24 | 9×11 | 6.97 |
| Commercial hybrids | |||||
| Kilima-ST | 5.09 | 6.51 | 6.48 | ||
| Kito-ST | 5.56 | 6.04 | 5.60 | ||
| Lishe-H1 | 5.14 | 6.12 | 6.74 | ||
| Lishe-K1 | 5.24 | 6.40 | 5.08 | ||
| Selian-H208 | 5.57 | 6.85 | 6.70 | ||
| Selian-H308 | 5.72 | 5.90 | 6.93 | ||
| Stuka-1 | 4.85 | 7.19 | 5.66 | ||
| TMV-1 | 6.18 | 6.17 | 5.53 | ||
| Vumilia-K1 | 5.43 | 6.20 | 5.29 | ||
| Statistics | |||||
| Grand mean | 3.03 | 4.39 | 4.38 | ||
| Lsd | 2.251 | 2.2048 | 1.4464 | ||
| Cv | 37.50 | 25.40 | 16.70 | ||
| Prob | <0.001 | <0.001 | <0.001 | ||
Where, 1=KS03-0B15-126, 2=EB04-0A01-304, 3=CML 159, 4=KS03-0B15-2, 5=KS03-0B15-45, 6=VL 05616, 7=KS03-0B15-47, 8=CML 395, 9=KS03-0B15-12, 10=CML 442 and 11=KL 05007-B-B.
Table 12: NLB disease severity of hybrids across three sites.
Discussion
Estimation of general combining ability (GCA) and specific combining ability (SCA)
The analysis model of (R2) explained 85, 64 and 73% effects for disease severity, yield and lesion numbers respectively to indicate that, the model was adequate. Level of significant differences among genotypes indicated by mean squares were also tested to justify the use of GCA and SCA and found to be highly significant (P<0.001) which fulfilled conditions suggested by Griffing [9]. Results revealed highly significant (P<0.001) environment mean squares for disease severity, yield and lesion number implying the significant impact of environment on genotypes. This could be caused by weather, edaphic factors and initial inoculum concentrations in the test sites which call for area specific cultivar recommendations as suggested by Macharia et al. [38].
Mean sum of squares of GCA was highly significant (P<0.001) on disease severity and lesion numbers and significant on yield (P<0.05) to denote the additive gene action expression. At the same time SCA mean squares were highly significant on disease severity and yield only to imply non-additive gene expression on those traits. These findings suggest that, both additive and non-additive gene effects are involved in the expression of NLB disease resistance in maize. Other researchers have reported on the additive and non-additive gene actions to express NLB disease resistance in maize [14,23,39,40]. The percentage of GCA contribution was 90.81% and 85.41% for disease severity and lesion numbers respectively. By considering levels of significant through mean squares and percentage of GCA contribution, it was obvious that additive gene action was more important than non-additive gene action for the expression of NLB disease resistance in maize germplasm which is in accordance to the findings of Sigulas et al. [25].
Parents combining ability on disease severity and yield
In disease resistance studies, negative GCA is desired while positive GCA effects are not desired. The GCA of nine parents had negative highly significant (P<0.001) effects on disease severity indicating additive gene effects for resistance. On yield, parent EB04-0A01-304 had significant positive GCA effects. Parents with positive GCA have the potential to impart high yielding characteristics to the next generation. Therefore, the combination of parents with negative GCA for disease severity and positive GCA for yield are likely to produce high yielding F1 hybrid.
Individual sites analysis also showed similar results for NLB disease severity inheritance. All parents had highly negative significant effect for NLB disease severity with the exception to CML 395 and KS03- 0B15-12. However, on yield the same CML 395 and KS03-0B15-12 parents had significant positive GCA (P<0.001) effects for yield. This means that these parents contributed to NLB disease susceptibility at the same time contributed to high yield potential. Based on individual site analysis, parents like CML 395 and KS03-0B15-12 need to be improved for NLB disease resistance so as to have parents with both high yielding and resistant potentials.
Specific combining ability (SCA) on NLB disease severity on F1 hybrids
The positive significant (P<0.005) SCA effects on disease severity showed by F1 hybrids indicates the non-additive gene action as expressed in CML 159 × CML 395 and VL 05616 × KS03-0B15-12 hybrids which could be probably due to masked susceptibility or maternal cytoplasmic effects of VL 05616 and CML 159 parents . However, the clear reason was not established by this study.
Reciprocal, maternal and non- maternal effects on yield
This study revealed that reciprocal effect which is associated with maternal and non-maternal effects were involved in yield expression. These gene actions have the tendency of reducing breeding efficiency through masking genetic variance [41]. The significant contribution of reciprocal, maternal and non-maternal effects on yield indicates the contribution of cytoplasmic genes and nuclear gene effects to bring an impact on maize yield. Some environmental factors like drought can enhance maternal effects as recorded by Derera et al. [42]. Thus, appropriate mating designs such as diallel cross and North Carolina design II can be employed to improve maize breeding procedures which encompass estimation of reciprocal effects depending on the number of parents to be used in the study [43].
Heterosis
Based on mid parent heterosis (MPH), F1 hybrids showed variations in the enhanced expression of disease severity and yield. The MPH was significantly higher for disease severity across sites to imply dominance gene action effects on the studied germplasm. Negative heterosis is desired in the studies of NLB disease resistance. All top ten experimental hybrids in each site had negative heterosis. On yield, a positive heterosis is desired. High positive heterosis on yield indicates the superiority performance for NLB disease reactions of F1 hybrids. The overall mid parent heterosis mean across sites was 151.84%. These results agree with Saleh et al. [37] who recorded much higher heterosis in maize.
Parents, commercial cultivars and F1 hybrid breeding potentials
The high variations in terms of NLB disease reactions and yield associated traits showed by parents implied high potential for breeding progress and yield increase in maize production. The selected inbred lines clearly expressed their traits across three diverse agro-ecological zones to show genetic potential for NLB disease reactions. The mean severity was higher at ARI Selian (22.9%) and ARI-Tumbi had the lowest (10.45%). The higher disease prevalence on parents and hybrids at ARI-Uyole and ARI-Selian could be attributed to weather conditions, land use and increased pathogen pathogenicity. Uyole is located in the highlands with favorable rainfall and temperature for disease development while Selian records more rainfall than Tumbi site. Uyole and Selian sites are characterized by land shortage which results in intensive land use with limited crop rotation flexibility. These practices result in accumulation of inoculums in maize stovers and alternative hosts. On the other hand ARI-Tumbi has less rainfall compared to the other two sites. At the same time, Tumbi site is characterized by abundant land to allow crop rotation, natural fallow, improved fallow and improved woodlots [44]. Frequent bush fire and free range animal grazing system which are common practices at Tumbi area could be other factors leading to relatively low level of NLB disease infestation in that area.
All commercial hybrids showed susceptibility to NLB disease with the scale of more than five. According to Ngugi et al. [45] and Reid [29], the detected commercial cultivar susceptibility can be classified as medium to high. Thus, all those commercial cultivars succumbed to NLB disease indicating their susceptibility. This implies that, farmers in Tanzania are growing NLB disease susceptible cultivars which could justify the disease resurgence and outbreaks in the near future if appropriate measures are not put in place. However, there was differential performance of individual hybrids to signify the importance of interactions which calls for area specific breeding procedures.
Conclusions
The following conclusions were drawn from this study
a. This study showed the predominance of additive gene action for controlling the NLB disease resistance in maize. In the study the mean sum of squares for GCA was highly significant (P<0.001) on disease severity indicating the predominance of additive gene action. This was further emphasized by the high GCA contribution for disease severity (91%) and lesion number (85%). The majority of parents had negative CGA to imply contribution to disease resistance on their progenies.
b. The mid parent heterosis for NLB disease severity ranged 93.46- 361.99%. Genotypes with negative heterosis to NLB disease is desired because they imply superiority performance of progenies towards resistance direction.
c. In this study reciprocal and maternal effects had nonsignificant (P>0.05) effects on the inheritance of the NLB disease severity.
d. Due to preponderance of the additive gene action, recurrent selection could be used to improve the resistance of inbreeds while the non-additive gene action could be exploited for breeding resistant hybrids in maize breeding program.
Acknowledgments
We are indebted to farmers in the study areas, The Alliance for a Green Revolution in Africa (AGRA) for financial support, the African Centre for Crop Improvement (ACCI) staff and management and the Ministry of Agriculture Food Security and Co-operatives for providing the enabling environment and study permission.
Discloser Statement
There is no potential conflict of interest reported by the authors in this article.
Funding
The financial support of the study was provided by The Alliance for a Green Revolution in Africa (AGRA) through the African Centre for Crop Improvement (ACCI) [grant number PASS88] at Kwazulu-Natal University, South Africa.
References
- CPD (2001) Crop protection compendium. 2001 ed. CAB International, Wallingford, UK.
- DeVries J, Toenniessen GH. (2001) Securing the harvest: biotechnology, breeding, and seed systems for African crops. CABI,New York
- Raymundo AD, Hooker AL(1981) Measuring the relationship between northern corn leaf blight and yield losses. Plant Disease 65:325-327.
- Payak MM, Renfro BL(1968) Combating maize disease. Indian Farmer Disease 1:53-58.
- Adipala E, Lipps PE, Madden LV(1993) Effect of disease assessment methods on predicting yield loss due to northern leaf blight of maize. African Crop Science Journal 2:167-178.
- Carson ML (2006) Response of a maize synthetic to select for components of partial resistance to Exserohilumturcicum. Plant Disease 90:910-914.
- Turner MT, Johnson ER(1980) Race of Helminthosporium not controlled by Ht genetic resistance in corn in the American corn belt. Plant Diseases 64:216-217.
- Christie BR, Shattuck VI. (1992) The diallel cross: design, analysis and use for plant breeders. Plant breeding reviews 9: 9-36.
- Griffing JB(1956) Concept of general and specific combining ability in relation to diallel crossing systems. Australian Journal of Biological Science 9:463-493.
- Gupta S, Kageyama S(1994)Optimal complete diallel crosses. Biometrika 81:420-424.
- Karaya H, Njoroge K, Mugo S, Nderitu H(2009) Combining ability among twenty insect resistant maize inbred lines resistant to Chilopartellus and Busseolafusca stem borers. International Journal of Plant Production 3:1735-8043.
- Lim SM,(1975) Diallel analysis for reactions of eight inbreds to Helminthosporiummaysdis race T. Phytopathology 65:10-15.
- Vivek BS, Odongo O, Njuguna J, Imanywoha J, Bigirwa G, et al. (2010) Diallel analysis of grain yield and resistance to seven diseases of 12 African maize (Zea mays L.) inbred lines. Euphytica 172:329-340.
- Vieira RA, Scapim CA, Moterle LM, Tessmann DJ, Conrado TV, et al. (2009) Diallel analysis of leaf disease resistance in inbred Brazilian popcorn cultivars. Genetics and Molecular Research 8:1427-1436.
- Bigirwa G, Julian AM, Adipala E (1993) Characterization of Ugandan isolates of Exserohilumturcicum from maize. African Crop Science Journal 1: 69-72.
- Ceballos H, Deutsch JA, Gutiérrez H(1991)Recurrent selection for resistance to Exserohilumturcicumin eight subtropical maize populations. Crop Science 31:964-971.
- Ferguson LM, Carson ML(2004)Spatial diversity of Setosphaeriaturcica sampled from the eastern United States. Phytopathology 94:892-900.
- Ferguson LM, Carson Ml(2007)Temporal variation in Setosphaeriaturcica between 1974 and 1994 and origin of Races 1, 23, and 23N in the United States. Phytopathology 97:1501-1511.
- Pratt RC, Gordon SG. (2006) Breeding for resistance to maize foliar pathogens. Plant Breeding Reviews 27: 119.
- Singh R, Mani VP, Koranga KS, Bisht GS, Khandelwal RS et al. (2004) Identification of additional sources of resistamce to Exserohilumturcicum in maize (Zea mays L.). SABRAO Journal of Breeding and Genetics 36: 45-47.
- Wisser RJ, Balint-Kurt BJ, Nelson RJ(2006)The genetic architecture of disease resistance in maize: A synthesis of published studies. Phytopathology 96:120-129.
- Pataky JK, Raid RN, Toit LJD, Schueneman TJ(1998) Disease severity and yield of sweet corn hybrids with resistance to northern leaf blight. Plant Disease 82:57-63.
- Sharma RC, Payak MM(1990) Durable resistance to two leaf blights in two maize inbred lines. TAG Theoretical and Applied Genetics 80:542-544.
- Ogliari JB, Guimarães MA, Geraldi IO, Camargo LEA(2005) New resistance genes in the Zea mays: Exserohilumturcicumpathosystem. Genetics and Molecular Biology 28:435-439.
- Hughes GR, Hooker AL(1971) Gene action conditioning resistance to northern leaf blight in maize. Crop Science 11:180-184.
- Sigulas KM, Hill RRJ, Ayers JE(1988) Genetic analysis of Exserohilumturcicum lesion expansion on corn. Phytopathology 78:149-153.
- Welz HG, Geiger HH (2000) Genes for resistance to northern corn leaf blight in diverse maize populations. Plant Breeding 119:1-14.
- Beyene Y, Mugo S, Gakunga J, Karaya H, Mutinda C, et al. (2011) Combining ability of maize (Zea mays L.) inbred lines resistant to stem borers. African Journal of Biotechnology 10:4759-4766.
- Muiru WM, Koopmann B, Tiedemann AV, Mutitu EW, Kimenju JW(2010) Race typing and evolution of aggressiveness of Exserohilumturcicum isolates of Kenyan, German and Australian origin. The World Journal of Agricultural Science 6:277-284.
- Reid LM, ZhuX (2005) Screening corn for resistance to common diseases in Canada Agriculture and agri-food Canada. Central Experimental Farm, Ottawa, Ontario, Canada.
- David O, Kempton RA, Nevison IM(1996) Designs for controlling inter-plot competition in variety trials. The Journal of Agricultural Science 127: 285-288.
- IBPGR (1991) Descriptors for maize. International Maize and Wheat Improvement Center, Mexico City, International Board for Plant Genetic Resources, Rome, Italy.
- Lindskog A, Moldanova J (1994) The influence of the origin, season and time of the day on the distribution of individual NMHC measured at Rörvik, Sweden. Atmospheric Environment 28: 2383-2398.
- Payne R, Murray D, Harding S, Baird D, Soutar D (2007) Genstat for Windows.10th ed. Introduction, VSN International, Hemel Hempstead, UK.
- Hariprasanna K, Lai C, Radhakrishnan T, Gor HK, Chikani BM (2008) Analysis of diallel cross for some physical-quality traits in peanut (Arachishypogaea L.).Euphytica 160: 49-57.
- Falconer DS, Mackay TFC(1996) Introduction to quantitative genetics. 4th ed.Printice Hall,Harlo, UK.
- Saleh GB, Abdullah D, Anuar AR(2002) Performance, heterosis and heritability in selected tropical maize single, double and three-way cross hybrids. Journal of Agricultural Science 138:21-28.
- Macharia CN, Njeru CM, Gichagi AW, Kamundia JW, Wasike VW, et al.(2009) Determination of location specific agronomic recommendations for a maize based farming system in western Kenya. African Crop Science Conference Proceedings 9: 221-226.
- Dingerdissen AL, Geiger HH, Lee M, Schechert A, Welz HG(1996) Internal mapping of genes for quantitative resistance of maize to Setosphaeriaturcica, cause of northern leaf blight, in a tropical environment. Molecular Breeding 2:143-156.
- Okello DK, Manna R, Imanywoha J, Pixley K, Edema R(2006) Agronomic performance and breeding potential of selected inbred lines for improvement of protein quality of adapted Ugandan maize germplasm. African Crop Science Journal 14:37-47.
- Durga KK, Reddy BVS,Reddy MSS, Ganesh M(2008) Influence of cytoplasm on the occurrence of leaf blight (Exserohilumturcicum) (Pass) in sorghum (Sorghum bicolorL.) Moench. Indian Journal of Agricultural Research 42: 97-101.
- Derera J, Tongoona P, Vivek B, Mark DL (2008) Gene actions controlling grain yield and secondary traits in southern Africa maize hybrids under drought and non-drought environment. Euphytica 162:411-422.
- Hallauer AR, Miranda JB(1988) Quantitative genetics in maize breeding. 2nd ed. Iowa State University, Iowa.
- Nyadzi GI, Janssen BH, Oenema O(2006) Analysis of the effects of rotational woodlots on the nutrition and yield of maize following trees in western Tanzania. Agriculture, Ecosystems & Environment 116:93-103.
- Ngugi HK, King SB, Abayo GO, Reddy YVR(2002) Prevalence, incidence, and severity of sorghum diseases in western Kenya. Plant Diseases 86:65-70.
Relevant Topics
- Agricultural science
- Agronomy
- Climate impact on crops
- Crop Productivity
- Crop Sciences
- Crop Technology
- Field Crops Research
- Hybrid Seed Technology
- Irrigation Technology
- Organic Cover Crops
- Organic Crops
- Pest Management
- Plant Genetics
- Plant Breeding
- Plant Nutrition
- Seed Production
- Seed Science and Technology
- Soil Fertility
- Weed Control
Recommended Journals
Article Tools
Article Usage
- Total views: 3918
- [From(publication date):
April-2017 - Jul 13, 2025] - Breakdown by view type
- HTML page views : 2986
- PDF downloads : 932
