Clinical Significance of Beclin-1 Dependent Autophagy Induced by Imatinib in Chronic Myeloid Leukemia Patients
Received: 15-Feb-2017 / Accepted Date: 26-Jun-2017 / Published Date: 30-Jun-2017
Abstract
Background and objectives: The introduction of imatinib as one of the tyrosine kinase inhibitors (TKI) has revolutionized the treatment of chronic myelogenous leukemia (CML), representing the first target specific drug. Autophagy is a constitutive homeostatic mechanism for intracellular recycling and metabolic regulation. The role of autophagy in cancer is very complex, it is tumor suppression mechanism yet it enables tumor cell survival in stress. The aim of the present study was to assess autophagy induced by imatinib therapy through beclin-1 gene in CML patients, and to evaluate beclin-1 gene expression in relation to patients’ clinical and hematological response.
Subjects and methods: The present study was carried out on 50 subjects, 35 newly diagnosed CML patients who were classified into 2 groups: (Group I) including 17 CML patients and (Group II) including 18 CML patients in accelerated phase, and 15 apparently healthy subjects served as controls (Group III). All patients were treated with imatinib and followed up after six weeks of treatment. Each patient was subjected to complete history taking, clinical examination along with laboratory investigations in the form of liver & renal function tests, random blood glucose, ESR, CBC & examination of Leishman-stained peripheral blood and Bone marrow aspiration and examination of Leishman-stained smears. The diagnosis was confirmed by detection of BCR-ABL gene expression by quantitative RT-PCR. Beclin-1 gene expression in peripheral blood samples was assessed as well via quantitative RT-PCR for all subjects along with its reassessment in follow-up samples for all patients.
Results: All patients in the study have achieved complete clinical and hematological response after six weeks of the beginning of imatinib therapy along with a highly significant increase in beclin-1 gene expression levels after treatment, (P<0.001) than before treatment in both patients groups I & II. There was no significant difference between group I & group II, regarding beclin-1 gene expression results after treatment (p=0.631). There was a high statistically significant negative correlation between beclin-1 gene expression before treatment and BCR-ABL gene expression results (r=-0.844 & -0.822) in group I and II respectively.
Conclusions: We concluded that beclin-1 dependent autophagy is increased after the beginning of imatinib therapy indicating induction of autophagy in CML patients, along with complete clinical and hematological response.
Keywords: Chronic myeloid leukemia; Imatinib; Autophagy
17313Introduction
Chronic myeloid leukemia (CML) is a myeloproliferative neoplasm, characterized by expansion of pluripotent bone marrow stem cells. The hallmark of the disease is the presence of a reciprocal t (9;22) (q34;q11.2), resulting in a BCR-ABL fusion gene. It was shown that this alteration generates a fusion between c-ABL (human homologue of the Abelson murine leukemia virus), a tyrosine kinase encoding oncogene, and BCR (breakpoint cluster region), the function of which is still not clear. The chimeric BCR-ABL protein possesses cellular-transforming ability that is ascribed to the elevated tyrosine kinase (TK) activity of the molecule. Further studies have established BCR-ABL as a leukemogenic oncogene, and both mouse models and in vitro assays have shown that BCR-ABL, as the sole oncogenic event, is sufficient to induce leukemia [1].
The introduction of imatinib as one of the tyrosine kinase inhibitors (TKI) in the treatment of chronic myeloid leukemia represent the most successful example of targeted therapy in human cancers [2]. Imatinib is a chemotherapeutic agent that specifically binds to the adenosine triphosphate binding pocket of the BCR/ABL fusion protein. This binding inhibits the subsequent phosphorylation events of the target proteins and suppresses cell proliferation [3].
Autophagy refers to the cellular degradation process in which cytoplasmic organelles are sequestered by autophagosome and then degraded inside lysosomes [4]. Autophagy can be induced by a number of stressors, acting to degrade protein polymers, oxidized lipids, injured organelles, as well as intracellular pathogens [5]. It is related with a series of neurodegenerative diseases, liver diseases, myopathy, tumor progression, aging, infection, immunization and inflammatory diseases [4]. The formation of autophagosome, including nucleation and expansion of the autophagosomal membrane, is dependent on the activity of ATG (autophagy-related) proteins, including the Beclin-1 complex, the ATG, and light chain [3] systems [6]. Autophagic degradation had long been regarded as bulk and nonselective, now it is known that autophagy is more useful as it can also selectively degrade various targets, including protein aggregates, damaged mitochondria, and even intracellular pathogens and this is called selective autophagy. In most cases, selectivity is determined by receptor proteins [7]. Autophagy is, observed in basal levels in almost all types of cells, as it is the regulated pathway for cells to recycle their components, which are degraded through activation of the lysosomal machinery upon induction of autophagy. Under starvation conditions, cells degrade some of their internal components via autophagy to derive monomers which will subsequently be used for the production of energy [8].
There are three known types of autophagy: macroautophagy, microautophagy, and chaperone-mediated autophagy. Macroautophagy is characterized by the encompassment of cellular components such as organelles and cytosolic protein aggregates by a double-membrane structure called autophagosome. This event can be initiated through the activation of serine/threonine kinase target of rapamycin (TOR) upon nutrient starvation [9].
ATG family of genes having more than 15 members is important in the initiation, expansion, and completion of the autophagosome. ATG5 (or APG5L) is required for the vesicle expansion and completion. ATG6 (or Beclin-1) is responsible for vesicle nucleation by forming the phosphatidylinositol 3-kinase complex along with Vps15 and Vps34 proteins [10].
Beclin-1 is expressed in many human and murine tissues and is localized primarily within cytoplasmic structures, including the endoplasmic reticulum, mitochondria and the perinuclear membrane [11]. Mutations of the Beclin-1 interfere with its abilities to promote nutrient deprivation-induced autophagy and suppress tumorigenesis 6.
Reduced beclin-1 gene expression was found in many cancers including esophageal, ovarian and lung cancer cells. On the other hand, there is increased expression in other cancers including colorectal and gastric cancers [12].
LC3 (or MAP1ALC3) is another key player required for the autophagosome expansion, followed by the fusion with the lysosome [13]. Then, hydrolases degrade its constituents and resultant monomers can be utilized by the cell [14]. Unlike macroautophagy, in microautophagy. Instead, lysosome engulfs targets to be degraded by invagination of the lysosomal membrane. This type of autophagy can also be initiated by mTOR signaling when nutrient in the medium is deprived [10]. The last known mechanism of the autophagy, chaperone-mediated autophagy, involves degradation of individual proteins [15].
The role of autophagy in health and disease is currently under the intensive scope of scientific research, as autophagy has been suggested to work as a double-edged sword in cancer development and progression by inducing both tumor cell survival and death. These diverse effects depend on to a large degree on the type of tumor, stage of disease, and nature of treatment.
BCR-ABL signaling mimics growth factor activation leading to inhibition of autophagy. Inhibition of BCR-ABL by TKIs has now been shown to not only induce apoptosis but also autophagy a similar effect to that seen after growth factor withdrawal [2] In line with this, it has been demonstrated that BCR-ABL–expressing mouse hemopoietic precursor cells have low basal levels of autophagy but are highly dependent on this process [2].
The aim of the present study was to assess autophagy induced by imatinib therapy through beclin-1 gene in CML patients. Also, our goal was involving the evaluation of autophagy induction by imatinib in relation to patients’ clinical and hematological response.
Subjects And Methods
The study population included 50 subjects, 35 newly diagnosed chronic myeloid leukemia patients who were selected from Minia and South Egypt Cancer Institutes, and fifteen apparently healthy subjects in the period from June 2014 to April 2016 and were followed up after six weeks after imatinib therapy. (Group I) included 17 patients including 13 males and 4 females, newly diagnosed with chronic myeloid leukemia, their ages ranged from 30 to 67 years. (Group II) included 18 CML patients in accelerated phase, (they were selected according to WHO criteria for accelerated phase of CML, all of them had BM blasts (10-19%) and four of them had a platelet count <100,000/microlitre), they were 11 males and 7 females, their ages ranged from 31 to 70 years. Group III (control group) included 15 apparently healthy subjects matched for age and sex for patient group, they were 10 males and 5 females and their ages ranged from 30 to 70 years.
Both patients and control groups were subjected to the following: complete history was taken, clinical examination and laboratory investigations: CBC determined by automated cell counter (Cell Dyn 3500, Abbott Diagnostics, USA), ESR determined by Westergren method, random blood glucose and liver function & renal function tests including (urea, creatinine and uric acid) analyzed by (Integra 400 plus, Roche Diagnostics, Germany). All Patients were presenting with hugely enlarged spleen and were diagnosed for CMLby: 1-Examination of Leishman-stained peripheral blood smears for differential leucocytic count and assessment of blast cell number, 2-Bone marrow aspiration by marrow puncture needles (Klima type) either from anterior or posterior superior iliac spine and examination of Leishman-stained smears. 3-BCR-ABL gene expression detection by quantitative RT-PCR (Light cycler® Roche, Germany). All subjects included were assessed for Beclin-1 gene expression detection by quantitative RT-PCR using (Light cycler® Roche, Germany) on peripheral blood sample withdrawn on a sterile K3EDTA tubes, before and after 6 weeks of imatinib treatment for all patients along with clinical and hematological re-evaluation.
Preparation of total RNA
Total RNA (including small RNAs as well as miRNAs) was extracted from EDTA whole blood samples using QIAamp RNA Blood Mini Kit Catalog no. 52304 supplied by (QIAGEN, Germany), according to the manufacturer's instructions.
Principle
QIAamp spin columns represent a technology for total RNA preparation that combines the selective binding properties of a silica– based membrane with the speed and convenience of microspin technology.
Reverse transcription (cDNA synthesis) and Beclin-1 gene expression
Template RNA was reverse transcribed to cDNA using the QuantiTect Reverse Transcription kit supplied by QIAGEN, Germany. The reverse-transcription master mix was prepared on ice by adding 1 μl Reverse-transcription master mix (Quantiscript Reverse Transcriptase), 4 μl Quantiscript RT Buffer, 1 μl RT Primer Mix and 14 μl Template RNA. Total reaction volume was 20 μl.
cDNA from all samples was used in the Two-Step RT-PCR with singleplex detection protocol, this protocol is for use with the QuantiFast Probe PCR+ROX Vial Kit and one QuantiFast Probe Assay. It was supplied by (Applied Biosystems, USA).
Procedure
Reaction mix was prepared by adding 12.5 μl 2 x QuantiFast Probe PCR Master Mix, 1.25 μl 20 x QuantiFast Probe Assay, 4.75 μl Template DNA or cDNA and 4.75 μl RNase-free water.
Beclin-1 forward and reverse primers
Beclin-1 forward primer 5`-CAAGATCCTGGACCGTGTCA-3` and beclin-1 reverse primer 5`-TGGCACTTCTGTGGACATCA-3` with the dual-labeled probes, TaqMan probes, contain a fluorescent reporter and a quencher at their 5' and 3' ends, respectively. The TaqMan probe sequence was CCGCACGGGACCAGAC. During the extension phase of PCR, the 5'-3' exonuclease activity of HotStarTaq plus DNA polymerase cleaves the fluorophore from the quencher. This results in detectable fluorescence that is proportional to the amount of accumulated PCR product.
Housekeeping gene ACTNB (β-actin)
Target nucleic acids can be quantified using either absolute quantification or relative quantification. Absolute quantification determines the absolute amount of a target (expressed as a copy number or concentration), whereas relative quantification determines the ratio between the amount of a target and the amount of a reference nucleic acid, usually a suitable housekeeping gene. This normalized value can then be used to compare, for example, differential gene expression in different samples. ACTNB (β-actin) forward primer 5′-CCAACCGCGAGAAGATGA-3′ and reverse primer 5′-CCAGAGGCGTACAGGGATAG-3′.
The reaction mix was mixed thoroughly and appropriate volumes are dispensed into PCR tubes and using real time Light cycler® Roche, Germany. The following thermal profile was used: a single cycle for 5 min at 90°C for initial activation step, 30 s at 95°C for denaturation, followed by 40 amplification cycles of 30 s at 95°C and 30 sec at 60°C each (annealing-extension step). Each reporter signal was measured against the internal reference dye (ROX) signal to normalize for non-PCR-related fluctuations between samples. The data were collected at the annealing step of each cycle and the threshold cycle (Ct) for each sample was calculated by determining the point at which the fluorescence exceeded the threshold limit.
Data analysis was performed as the following
The range for target gene relative to a calibrator housekeeping gene (fold change in the target gene relative to the ACTNB endogenous control gene) is determined by Relative expression or Quantitation (Q)=2ΔΔCT (Cycle threshold)
ΔCT (CT Target–CT reference (housekeeping gene)
Δ(ΔCT)=ΔCT test sample–ΔCT calibrator sample.
Statistical analysis
Data were analyzed by using SPSS (Statistical Package for Social Science) version 16. Results were expressed as mean ± standard deviation (SD). Student t-test of Mann Whitney was used to compare variables between the two groups. One way ANOVA test for comparison between the three groups. Pearson correlation was used to perform correlation between different variables. Results were considered significant if P-value was <0.05 and highly significant if P-value<0.001.
Results
The present study included 35 CML patients and 15 apparently healthy volunteers as a control group. Demographic data of the studied groups are shown in (Table 1).
| Group I (Chronic) | Group II | Group III | P value | |||
|---|---|---|---|---|---|---|
| Age (years) | 0.975 | |||||
| Range | (30-67) | (31-70) | (31-70) | I vs. II | I vs. III | II vs. III |
| Mean ± SD | 50.5 ± 11.9 | 50.3 ± 10.2 | 49.7 ± 11.2 | 0.998 | 0.974 | 0.986 |
| Sex | 0.617 | |||||
| Male | 13 (76.5%) | 11 (61.1%) | 10 (66.7%) | I vs. II | I vs. III | II vs. III |
| Female | 4 (23.5%) | 7 (38.9%) | 5 (33.3%) | 0.328 | 0.538 | 0.741 |
Table 1: Demographic data of the studied groups.
Beclin-1 gene expression levels and patient clinical and hematological response: Regarding the patients’s clinical response, all patients showed impalpable spleen after six weeks with imatinib treatment. Beclin-1 gene expression level, total leucocytic count (TLC), hemoglobin (Hb) level and platelets count before and after treatment in group I (Table 2). Beclin-1 gene expression showed statistically significant high increase results after treatment, (P=<0.001) than before treatment.
| Group I (Chronic phase) (n=17) | Before treatment | After treatment | P value |
|---|---|---|---|
| Beclin-1 | |||
| Range | (0.16-115.8) | (20-200) | <0.001** |
| Mean ± SD | 45.0 ± 47.4 | 93.6 ± 65.4 | |
| TLC ( × 10³ cell/μl) | |||
| Range | (25-614) | (4.7-11) | <0.001** |
| Mean ± SD | 162.9 ± 157.3 | 8.1 ± 2.1 | |
| Hb (g/dl) | |||
| Range | (7.4-13) | (10.5-14.2) | 0.068 |
| Mean ± SD | 10.5 ± 1.7 | 11.9 ± 1.2 | |
| Platelet count ( × 10³/μl) | |||
| Range | (134-1000) | (145-409) | 0.003* |
| Mean ± SD | 506.4 ± 241.4 | 284.4 ± 91.0 | |
| Hb, haemoglobin; TLC, total leucocytic count. * P value is significant (when P<0.05), ** P value is highly significant (when P<0.001) | |||
Table 2: Beclin-1 gene expression, total leucocytic count, hemoglobin level and platelet count before and after treatment in group I.
There was high statistically significant decrease in TLC when comparing results before and after treatment, (P=<0.001), also there was statistically significant decrease in platelet count, (P value=0.003), while, there was no statistically significant difference between Hb level before and after treatment, (P=0.068). Beclin-1 gene expression level, total leucocytic count (TLC), hemoglobin (Hb) level and platelets count before and after treatment in group II (Table 3). Beclin-1 gene expression showed statistically significant high increase results after treatment (P=<0.001). There was high statistically significant decrease in TLC when comparing results before and after treatment, (P=<0.001), also, there was statistically significant increase in Hb level after treatment, (P=0.027), while, there was no statistically significant difference between platelet count before and after treatment, (P=0.102).
| Group II (Accelerated phase) (n=18) | Before treatment | After treatment | P value |
|---|---|---|---|
| Beclin-1 | |||
| Range | (0.03-118.7) | (10-180) | <0.001** |
| Mean ± SD | 38.7 ± 49.4 | 84.1 ± 61.7 | |
| TLC ( × 103 cell/μl) | |||
| Range | (36-843) | (4.6-10) | <0.001** |
| Mean ± SD | 271.1 ± 249.9 | 7.5 ± 1.8 | |
| Hb (g/dl) | |||
| Range | (7.2-13.5) | (10.8-15) | 0.027* |
| Mean ± SD | 10.8 ± 1.5 | 12.5 ± 1.0 | |
| Platelet count ( × 103/μl) | |||
| Range | (43-670) | (170-420) | 0.102 |
| Mean ± SD | 327.9 ± 186.7 | 249.1 ± 83.4 | |
| Hb: Haemoglobin; TLC: Total Leucocytic Count. * P value is significant (when P<0.05) ** P value is highly significant (when p- value<0.001). | |||
Table 3: Beclin-1 gene expression, total leucocytic count, hemoglobin level and platelets count before and after treatment in group II.
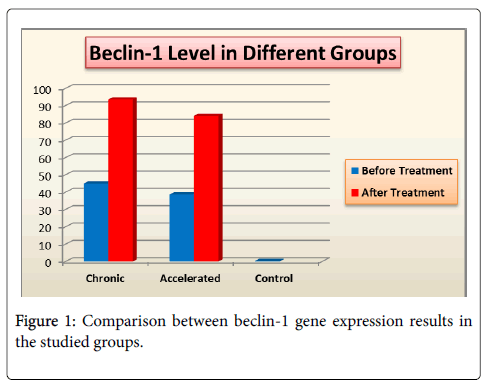
Beclin-1 gene expression levels in studied groups before and after treatment: Comparison between beclin-1 gene expressions in the studied groups (Figure 1). There was high statistically significant increase in beclin-1 gene expression results before treatment in group I when compared with group III (P=0.001) & moderate statistically significant increase in its results before treatment in group II when compared with group III (P=0.006) beclin-1 gene expression results after treatment, showed no statistically significant difference between group I & group II. Correlations between beclin-1 gene expression before treatment and BCR-ABL gene expression: Results of group I & II (Figures 2 and 3).
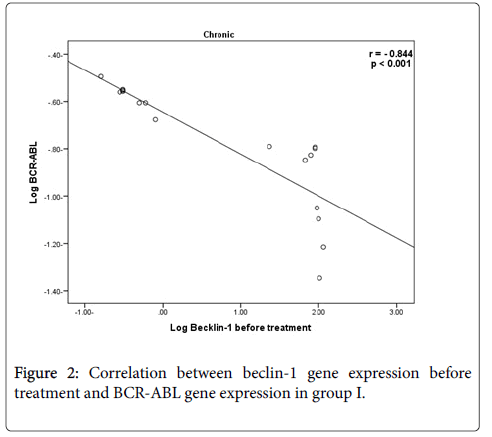
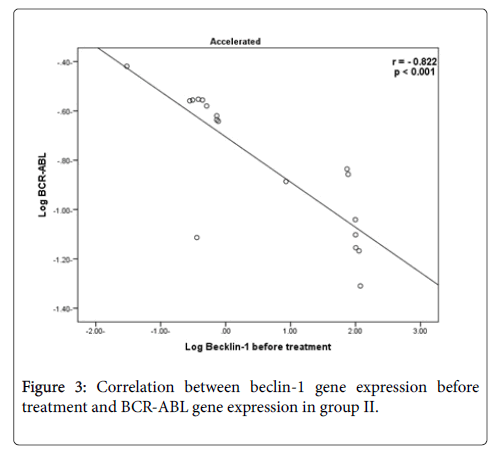
There was a high statistically significant negative correlation between beclin-1 gene expression before treatment and BCR-ABL gene results (r=-0.844 & -0.822) in group I and II respectively.
Discussion
Advances in chronic myeloid leukemia treatment, particularly regarding tyrosine kinase inhibitors, have introduced and recommend Imatinib, a breakpoint cluster region-Abelson murine leukemia tyrosine kinase inhibitor, which represents the most successful example of targeted therapy in human cancers 1.
Autophagy is a recycling process that leads to sequestration and degradation of damaged proteins and other intracellular material within lysosomes [16]. Although it is established that autophagy has a dual role in cancer, the dynamics by which this catabolic process suppresses or promotes cancer still remain complex. Efforts to reveal molecular and cellular mechanisms that might modulate autophagy activity in cancer cells have started to emerge [17]. Autophagy leads to tumor suppression and type-II programmed death of cancer cells, defense against bacterial or viral infection, and prevention of neurodegeneration via degrading toxic protein aggregates. But, autophagy might also contribute to tumor cell survival under nutrient-deprived conditions or by protection against cancer treatments [18].
In the present study 35 newly diagnosed CML patients were assessed for autophagy through beclin-1 gene expression detection in peripheral blood, then reassessment was done after the beginning of therapy with imatinib. All patients in this study (group I & II) developed complete clinical response with impalpable spleen as well as complete hematologic response, which was assessed by peripheral blood counts, peripheral blood smears and clinical evaluation, while Medhi et al. [19] reported that only 95% of patients developed complete hematologic response.
In the current study, there was highly significant negative correlation between beclin-1 gene expression before treatment in group I & II and BCR-ABL gene expression results, that was in accordance with Vignir et al., who reported that BCR-ABL signaling leads to activation of the PI3K/AKT pathway and mTOR with subsequent decreasing level of autophagy.
In all patients of the present study (group I & II), there was a highly significant increase in beclin-1 gene expression detection after the beginning of the treatment with imatinib which indicates induction of autophagy. That is in agreement with Geylani et al. [18], who demonstrated that there were dose-dependent increases in expression levels of ATG5 and beclin-1 genes in K562 cells exposed to 0.1, 1, and 10 nM imatinib, respectively. Also, this is consistent with Vignir et al. [2], who reported that inhibition of BCR-ABL by TKIs has now been shown not only to induce apoptosis but also autophagy, a similar effect to that seen after growth factor withdrawal. In agreement with the present study, Calabretta and Salomoni, [20] demonstrated that imatinib as one of the TKIs induces autophagy and that combining imatinib with Chloroquine (CQ), as an autophagy inhibitor, can potentiate the effect of TKI and restoring the sensitivity to imatinib.
The presence of leukemic stem cells which are insensitive to TKIs and contribute to the persistence of disease by representing a reservoir of self-renewing cells that replenish the disease after drug discontinuation. Moreover, given that BCR-ABL–expressing stem cells can survive without BCR-ABL signaling, therefore, alternative survival pathways responsible for leukemic stem cell survival after oncogene inactivation must be examined, one example of such a survival pathway is autophagy. These findings have refocused the interest of scientists towards drug combinations treating CML with TKIs and simultaneously targeting alternative survival mechanism as the autophagy mechanism [2].
Autophagy induced as result of imatinib treatment in the current study might be either due to cell death or it might protect the cells against the stress of chemotherapy. Based on our findings, we can suggest, that if autophagy is acting as a cell death mechanism besides the imatinib-dependent apoptosis, then, autophagy pathway might be promoted for more successful CML therapies. On the contrary, if autophagy acts as a protective mechanism for the cancer cell, in such cases, drugs blocking autophagy pathway might be incorporated into the therapy along with imatinib therapy. This approach might be useful in treating the imatinib-resistant CML.
Our study was limited by several factors: (1) Beclin-1 and BCR-ABL genes expression levels were not assessed after 3, 6, and 12 months of imatinib therapy {standardized response assessment is with real quantitative polymerase chain reaction and/or cytogenetics at 3, 6, and 12 months} which could provide important information about the relation between autophagy and the development of resistance to imatinib therapy. (2) The number of subjects that is small due to financial issues (3) Larger sample sizes and follow-up studies are required in future research to further expose Beclin-1 gene as a potential target of therapy if blocking autophagy pathway is desired, and reveal the effectiveness of using combinations between imatinib and chloroquine or any other autophagy inhibitors in increasing the cure rate for CML patients.
Conclusion
We concluded that beclin-1 dependent autophagy is increased after the beginning of imatinib therapy indicating induction of autophagy in CML patients, along with complete clinical and hematological response.
Ethical Considerations
The study was performed in accordance with the World Medical Association’s Declaration of Helsinki. It was approved by the research ethics committee of El-Minia University. The purpose and nature of the study were fully explained to the subjects and all of them signed written, informed consents before their enrollment in the study. All data were kept private and used for research purposes only.
References
- Philip A, Hagop M, Jorge E (2015) Diagnosis and Treatment of Chronic Myeloid Leukemia in Mayo Clin Proc 90: 1440-1454.
- Â Vignir H, Maria K, Tessa L, Holyoake (2011) Kill one bird with two stones: potential efficacy of BCR-ABL and autophagy inhibition in CML. Blood 118: 2035-2043.
- Chuanjiang Yu, Sivahari P, Tony M, Lena L, Zhenyu Y, et al. (2015) Beclin-1 Phosphorylation By BCR-ABL Is Crucial for CML Leukemogenesis By Suppression of Autophagy. Blood 126: 163-170.
- Yuan Zhou, Ying Li, Wei Jiang, Lin Zhou (2015) MAPK/JNK signaling: a potential autophagy regulation pathway. Biosci Rep 35: 199-207.
- Rabinowitz J, White E (2010) Autophagy and metabolism. Science 330: 1344–1348.
- Xiaoqian H, Bin Y, Xingshan L, Huixiang Y, Xishuang L (2015) Expression of Beclin1 in the colonic mucosa tissues of patients with ulcerative colitis. Int J Clin Exp Med 8: 21098–21105.
- Hitoshi N, Yoshinori O (2014) Autophagy: close contact keeps out the uninvited. Current Biology 24: 560-562.
- Shintani T, Klionsky DJ (2004) Autophagy in health and disease: a double-edged sword. Science 306: 990–995.
- Pattingre S, Espert L, Biard-Piechaczyk M, Codogno P (2008) Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie 90: 313–323.
- Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y (2008) Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell 19: 2039–2050.
- Kang R, Tang D, Lotze MT, Zeh HJ (2011) Apoptosis to autophagy switch triggered by the MHC class III-encoded receptor for advanced glycation endproducts (RAGE) Autophagy 7: 91–93.
- Kotsafti, Farnati, Cardin (2012) Autophagy and apoptosis-related genes in chronic liver disease and hepatocellular carcinoma. BMC Gastroenterology 12: 118-126.
- He H, Dang Y, Dai F, Guo Z, Wu J, et al. (2003) Posttranslational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem 278: 29278–29287.
- Luzio JP, Pryor PR, Bright NA (2007) Lysosomes: fusion and function. Nat Rev Mol Cell Biol 8: 622–632.
- Mijaljica D, Prescott M, Klionsky DJ, Devenish RJ (2007) Autophagy and vacuole homeostasis: a case for self-degradation? Autophagy 3: 417–421.
- Mizushima N (2007) Autophagy: process and function. Genes Dev 21: 2861-2873.
- Abibatou, Ashani (2016) Autophagy- An emerging target for melanoma therapy. F1000 Faculty Rev 1: 1888-1895.
- Geylani C, Huseyin A, Yusuf B ( 2011) Imatinib induces autophagy through BECLIN-1 and ATG5 genes in chronic myeloid leukemia cells. Hematology 16: 95-99.
- Medhi K, Raina V, Kumar L (2010) Response assessment of patients with chronic myeloid leukemia receiving imatinab mesylate therapy Leuk Lymphoma 51: 1850-185S4.
- Calabretta B, Salomoni P (2012) Suppression of autophagy by BCR/ABL. Front biosci 4: 453-460.
Citation: El-Sharkawy EA, Salah KM, Kamal Eldin AM, Ghobrial AG, Abdel-Hamid WM, et al. (2017) Clinical Significance of Beclin-1 Dependent Autophagy Induced by Imatinib in Chronic Myeloid Leukemia Patients. J Mucosal Immunol Res 1:103.
Copyright: ©2017 El-sharkawy EA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 3881
- [From(publication date): 0-2017 - Apr 03, 2025]
- Breakdown by view type
- HTML page views: 2981
- PDF downloads: 900