Research Article Open Access
Chitin Elicitor-Responsive Photon Emission is potentiated by Plant Activators through Priming of Salicylic Acid Signaling via OsWRKY45 in Rice
Hiroyuki Iyozumi*, Hideki Nukui and Kimihiko Kato
Shizuoka Prefectural Research Institute of Agriculture and Forestry, Tomigaoka, Iwata, Shizuoka 438-0803, Japan
- *Corresponding Author:
- Hiroyuki Iyozumi
Shizuoka Prefectural Research Institute of Agriculture and Forestry
Tomigaoka, Iwata
Shizuoka 438-0803, Japan
Tel: +81 538 36 1556
Fax: +81 538 37 8466
E-mail: hiroyuki1_iyozumi@pref.shizuoka.lg.jp
Received date: July 15, 2016; Accepted date: September 09, 2016; Published date: September 16, 2016
Citation: Iyozumi H, Nukui H, Kato K (2016) Chitin Elicitor-Responsive Photon Emission is potentiated by Plant Activators through Priming of Salicylic Acid Signaling via OsWRKY45 in Rice. J Rice Res 4:175. doi:10.4172/2375-4338.1000175
Copyright: © 2016 Iyozumi H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Rice Research: Open Access
Abstract
Priming of plant cells for faster and enhanced defense responses against pathogen attacks is a common feature of chemically or biologically induced resistance. The authors previously developed a priming detection system that detects priming as potentiation of chitin elicitor-responsive photon emission (C-ERPE) in rice cells pretreated with various types of chemical inducers of disease resistance, called plant activators. To elucidate the mechanisms underlying C-ERPE potentiation, the authors performed gene knockdown of OsWRKY45, a major regulator of salicylic acid (SA)-dependent defense responses in rice, and estimated the effects of SA isomers on C-ERPE potentiation. Plant activators induced a 200-300% increase in C-ERPE in the wild type, whereas OsWRKY45 knockdown attenuated the increase in C-ERPE to less than 60%. Native SA induced more than a 150% increase in C-ERPE, but structural isomers of SA were less effective (10-24% increase). These SA signaling-disruption experiments indicate that the potentiation of C-ERPE requires intrinsic components of hormonal signaling for defense, at least for priming by inducers of systemic acquired resistance.
Keywords
Priming; Plant activator; Rice; WRKY; Ultraweak photon emission; Elicitor
Abbreviations
UPE: Ultraweak Photon Emission; ERPE: Elicitor- Responsive Photon Emission; IR: Induced Resistance; SA: Salicylic Acid; SAR: Systemic Acquired Resistance; RNAi: RNA interference; PCR: Polymerase Chain Reaction; WT: Wild Type; RT-PCR: Reverse Transcription PCR; ASM: Acibenzolar-S-Methyl; TDL: Tiadinil; 3HBA: 3-Hydroxy Benzoic Acid; 4HBA: 4-Hydroxybenzoic Acid; PA: Phosphatidic Acid; ROS: Reactive Oxygen Species; PLD: Phospho Lipase D
Introduction
Rice is a major food staple throughout the world, especially in the Asia-Pacific region, parts of South and Central America and, increasingly, in Africa. It plays a pivotal role in the food security of over half of the world’s population [1,2]. In the past half-century, despite competition for water and land use with other crops or nonagricultural purposes, rice production has continued growing around the world [3-5]. In the past few decades, especially after the mapping and sequencing of its whole genome, rice has become an important target of Omics Research and other new technologies [6-8].
Ultraweak photon emission (UPE) from plant cells is a real-time indicator of plant defense responses and can be assessed noninvasively [9,10]. The authors discovered UPEs in leaf segments and cultured cells of rice treated with microbial (fungal) elicitors including chitin [11-14] and inorganic substances such as dipotassium hydrogen phosphate or copper chloride [15], and named them elicitor-responsive photon emissions (ERPEs). Other studies of bacteria and viruses have reported UPEs in plants that are induced by pathogen-derived molecules [16,17], and ERPE is thought to be common in plant-pathogen interactions.
Induced resistance (IR) of plants against pathogens is of interest in the study of crop protection, and chemicals that induce resistance in plants, so-called “plant activators”, have been investigated [18,19]. The selection of candidates for plant activators by detecting IR-related genes, so-called pathogenesis-related genes, has been reported using microarray analysis [20] or reporter gene assays [21-23] to accelerate plant activator screening. Faster and enhanced defense responses to pathogens or elicitors are characteristic cellular events in plants treated with plant activators; this effect is called “priming” [24,25]. Priming is common in types of IR mediated by various kinds of defense-related hormones, and the key factors involved in signaling have been discovered gradually [26]. In rice, the crucial roles of the transcription factor WRKY45 (OsWRKY45) in salicylic acid (SA)-mediated systemic acquired resistance (SAR) have been reported [27-29].
Although the current knowledge-based plant activator screening process is powerful when used in the context of “Omics” databases, there may be unknown mechanisms underlying IR. In addition, the current screening methods can require invasive processes in plants, which may affect the screening results.
To address these problems, considering the previous studies of IR and ERPEs, the authors proposed a simple method for the screening of plant activators, in which enhancement of ERPE by priming could be detected [30]. This method can detect the priming effects of various kinds of plant activators or defense-related hormones [30] and may be useful for preliminary and inclusive screening of chemical libraries for plant activators. Although the mechanisms underlying ERPEs have been revealed gradually [12-14], the mechanism through which priming potentiates ERPEs has not been clarified.
Here, the authors report that the priming for enhanced C-ERPE by pretreatment of rice cells with SAR inducers requires intrinsic SA signaling components, including the transcription factor OsWRKY45 . The authors also discuss the properties of C-ERPE in rice cells as a priming detector.
Materials and Methods
Plant materials and chemicals
Cell culture: Rice cells (Oryza sativa L. cv. Kimmaze) were maintained with shaking in modified N6 liquid medium [31] containing 1 mg/L of 2,4-dichlorophenoxyacetic acid (2,4-D) at pH 5.8, and 25°C in the dark with natural ventilation. Aliquots of 15 g (fresh weight) of cells were transferred to fresh medium every 10 days. Before plant activator and elicitor treatments, 1 mL portions of 10-day cultured cell suspensions containing 0.5 g of cells were dispensed onto plastic Petri dishes.
OsWRKY45 gene expression knockdown by RNA interference (RNAi): The OsWRKY45 (AK066225) RNAi vector was constructed using the Gateway® pENTR/D-TOPO Cloning Kit (Thermo Fisher Scientific, Waltham, MA, USA) and a pANDA vector as described [32,33]. The pANDA vector was kindly provided by Nara Institute of Science and Technology, Japan. Briefly, the 309 bp cDNA fragment of the 3′-untranslated region of OsWRKY45 mRNA (5′- GGACACGGGCCGGGTAAAACGATCGAAAGAAG ATGGATTCCACGCGTGTGTACAGAAATAATTAGCGGCAGCGC GGATCTTAATTTGGAACTTGCAAAGATACTCCTAATTAGCCTG GCTAGATTAGTTTGTAAATTCCTTGTTGATGTGTCGTCTCAGC TTTAAGCTGCAGACATGCTAGCAAGTAACAACACGATTAGTAC GTAGTAATGTGGTTCTTGATTATGAGCTGGGGGTCTTAACCTT TTTTGTGTGACAAGCAAGAGAAGAGGATTTGGGTACAATGTA ATCCTGTTCTTCCGCTTTCGA-3′) was amplified by polymerase chain reaction (PCR) using a primer set for directional cloning (forward 5′- CACCGGACACGGGCCGGGTAAAACGATCGAAAGA-3′ and reverse 5′-TCGAAAGCGGAAGAACAGGATTACATTGTACCCA-3′). The recombination reaction of the PCR product with the pENTR/DTOPO vector was performed according to the manufacturer’s instructions. The pANDA-OsWRKY45 -RNAi vector was constructed by incubating the entry clone (pENTR-OsWRKY45 ), which had been pretreated with the restriction enzyme NruI , and the pANDA vector with the Gateway LR Clonase Enzyme Mix (Thermo Fisher Scientific). Rice cells (cv. Kimmaze as the wild type, WT) were transformed with the pANDA-OsWRKY45 -RNAi vector (Supplemental Figure 1) by Agrobacterium -mediated transformation [34]. The transformed cell lines were selected in modified N6 liquid medium [31] containing carbenicillin (300 mg/L) and hygromycin (50 mg/L) while shaking at 100 rpm at pH 5.8, 25°C in the dark for 12 days with natural ventilation. Selection was repeated at least four times by transferring the selected lines to new medium every 12 days. In these lines, the gus linker sequence (Supplemental Figure 1) was detected by reverse transcription PCR (RT-PCR) to ascertain the triggering of doublestrand RNA production [32,33]. Ten-day-old cells were used for photon counting experiments and RNA extraction.
Plant activators and isomers: Acibenzolar-S-methyl (ASM, synonym; benzothiadiazole: BTH), tiadinil (TDL), SA, and its inactive isomers 3-hydroxy benzoic acid (3HBA) and 4-hydroxybenzoic acid (4HBA) were purchased from Wako Pure Chemicals (Osaka, Japan). Each chemical was dissolved in the solvent (98% v/v of N, Ndimethylformamide (Wako Pure Chemicals) and 2% (v/v) of Tween 20 (Wako Pure Chemicals)) and used as 100-fold concentrated stocks (20 mM). Aliquots of 1% (v/v) of the stock solutions were added to cell suspensions to adjust the plant activator concentrations to 200 μM. An equal volume of the solvent solution was used as a control treatment.
Elicitor experiments: N -acetylchitohexaose (Seikagaku Corporation, Tokyo, Japan) was used as a chitin elicitor of ERPE. N - acetylchitohexaose was dissolved in distilled water and adjusted to 20 μM as a 20-fold stock solution. Aliquots of 5% (v/v) of the stock solution were added to the rice cell suspensions to adjust the elicitor concentrations to 1 μM after the plant activator treatments. Equal volumes of distilled water were used as control treatments.
Photon counting experiments: UPE measurements were performed using a PCX-100 photon counter (Hamamatsu Photonics K.K., Hamamatsu City, Japan) as described [11]. Briefly, 10-day cultured rice cells were dispensed into 60 mm diameter plastic Petri dishes (Eiken Chemical Co. Ltd., Tokyo, Japan) and treated with each plant activator solution. The Petri dishes with cells were set in the light-tight box of the photon counter and photon counting was started. After pretreatment for 2 h, the photon counting was paused, a portion of elicitor solution was added to each dish, and the photon counting was continued. From the initial sample placement, all steps were performed in the dark to avoid external light exposure. All experiments were performed in triplicate at 26°C in an air-conditioned dark room. ERPE levels are expressed as 5 h integrated photon counts after chitin elicitor treatment and were calculated by subtracting the values for the watertreated controls (Supplemental Figure 2).
Quantitative RT-PCR analysis: Two hours after pretreatment with plant activators, 50 mg aliquots of cells were collected, frozen immediately in liquid nitrogen, and powdered using a mortar and pestle. Total RNA was extracted using RNeasy Plant Mini Kits (Qiagen, Hilden, Germany). Complementary DNA was obtained from 500 ng of total RNA using Quantitect® Reverse Transcription Kits with a genome DNA eraser (Thermo Fisher Scientific). Quantitative PCR reactions were performed using SYBR® Premix Ex Taq (TaKaRa Bio Inc., Otsu, Japan) in an Mx3000-P Q PCR system (Agilent Technologies, Palo Alto, CA, USA). The cycling conditions were 10 s of polymerase activation at 95°C followed by 40 cycles at 95°C for 5 s and 64°C for 30 s. The expression level of OsWRKY45 was normalized against the expression of the Rice Ubiquitin 1 gene (Ubq1 ) in the same sample. The primers used were forward 5′-GAACGACGAGGTTGTCTTCG-3′ and reverse 5′-ACGCGTGGAATCCATCTTCT-3′ for OsWRKY45 ; and forward 5′-CCAGTAAGTCCTCAGCCATGGAG-3′ and reverse 5′-GGACACAATGATTAGGGATCACTT-3′ for Ubq1 .
Results and Discussion
Effects of OsWRKY45 knockdown on the priming of rice cells by SAR inducers for enhanced ERPE
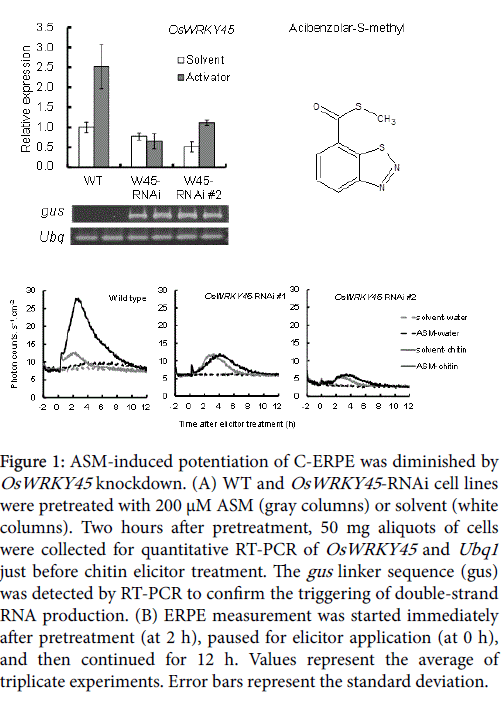
The OsWRKY45 -RNAi-treated cell lines were similar to WT cells in appearance (Supplemental Figure 2) and in Ubq1 expression level of (Figure 1A). OsWRKY45 gene expression in the solvent-pretreated OsWRKY45 -RNAi cell line were almost equal to or lowers than that in the solvent-pretreated WT line (Figure 1A, white columns). The average expression rate compared with the WT was 77.6% in OsWRKY45 -RNAi line #1 and 51.2% in OsWRKY45 -RNAi line #2. ASM induces SAR in monocots and dicots [35,36], and is a powerful inducer of OsWRKY45 transcription factor expression in rice [27]. However, OsWRKY45 expression was lower in the ASM-pretreated OsWRKY45 -RNAi line than in the WT (Figure 1A, gray columns). The average expression rate compared with the WT was 25.9% in OsWRKY45 -RNAi line #1 and 44.3% in OsWRKY45 -RNAi line #2. This confirmed the knockdown of OSWRKY45 .
Figure 1: ASM-induced potentiation of C-ERPE was diminished by OsWRKY45 knockdown. (A) WT and OsWRKY45 -RNAi cell lines were pretreated with 200 μM ASM (gray columns) or solvent (white columns). Two hours after pretreatment, 50 mg aliquots of cells were collected for quantitative RT-PCR of OsWRKY45 and Ubq1 just before chitin elicitor treatment. The gus linker sequence (gus) was detected by RT-PCR to confirm the triggering of double-strand RNA production. (B) ERPE measurement was started immediately after pretreatment (at 2 h), paused for elicitor application (at 0 h), and then continued for 12 h. Values represent the average of triplicate experiments. Error bars represent the standard deviation.
Treatment with 1 μM chitin elicitor induces biphasic ERPE in rice cells, which peaks a few minutes and 2 h after treatment [12]. The CERPE was nearly similar to that of the WT in OsWRKY45 -RNAi cell line #1 but was weaker in OsWRKY45 -RNAi cell line #2. In these two cell lines, the second peak appeared 1 h later than that of the WT cells (Figure 1B, gray line). The effect of OsWRKY45 -RNAi was more apparent during the potentiation of ERPE by pretreatment of rice cells with ASM. In WT cells, the 5 h integrated C-ERPE count increased after ASM pretreatment by 339% compared to that after solvent pretreatment (Figure 1B, black line). By contrast, the increase rate in C-ERPE by ASM pretreatment was suppressed in the OsWRKY45 - RNAi lines (Figure 1B; -3% in OsWRKY45 -RNAi line #1 and 57% in OsWRKY45 -RNAi line #2). Taken together, these experiments confirmed the contribution of OsWRKY45 to ERPE potentiation by ASM.
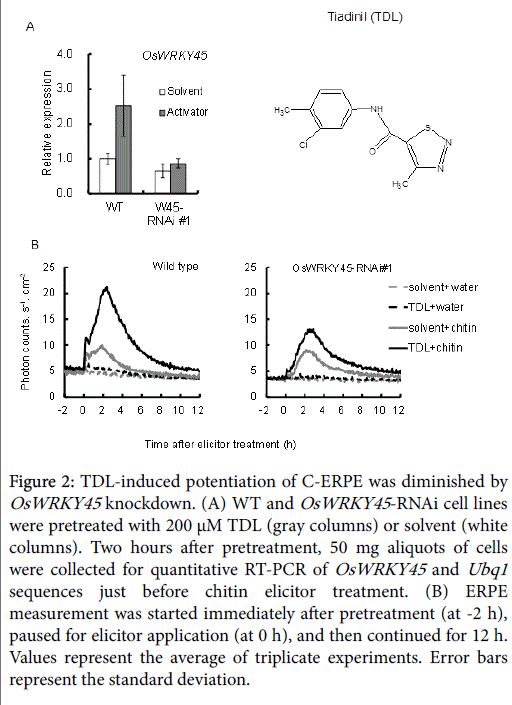
TDL, which has been commercialized as a plant activator for rice disease, affects defense signaling in a similar way to ASM in tobacco [37,38]. In rice cells, TDL also induced OsWRKY45 expression in WT cells, and this was suppressed in the RNAi line (Figure 2A, gray columns). The average expression rate compared with WT cells was 34.3% in OsWRKY45 -RNAi line #1). The 5 h integrated C-ERPE count increased after TDL pretreatment by 228% compared to that after solvent pretreatment in WT cells, and a 78% increase was observed in OsWRKY45 RNAi line #1 (Figure 2B). This confirmed the contribution of OsWRKY45 to ERPE potentiation by TDL.
Figure 2: TDL-induced potentiation of C-ERPE was diminished by OsWRKY45 knockdown. (A) WT and OsWRKY45 -RNAi cell lines were pretreated with 200 μM TDL (gray columns) or solvent (white columns). Two hours after pretreatment, 50 mg aliquots of cells were collected for quantitative RT-PCR of OsWRKY45 and Ubq1 sequences just before chitin elicitor treatment. (B) ERPE measurement was started immediately after pretreatment (at -2 h), paused for elicitor application (at 0 h), and then continued for 12 h. Values represent the average of triplicate experiments. Error bars represent the standard deviation.
Contribution of SA signaling via OsWRKY45 to the potentiation of C-ERPE by priming
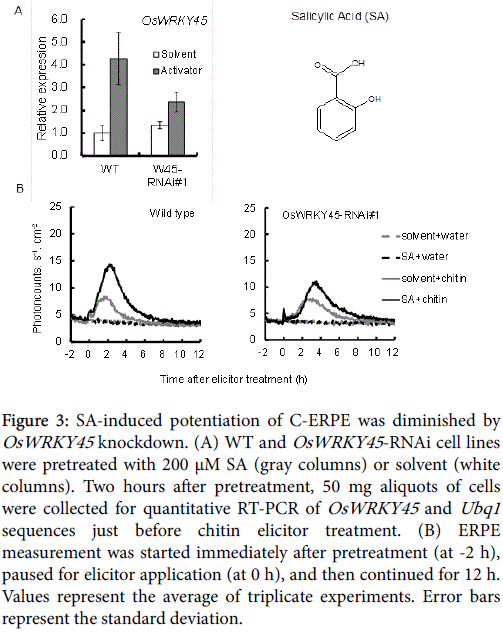
ASM and TDL are functional analogues of SA, and induce SAR by affecting the downstream events of SA-mediated defense signaling cascade in dicots [36,37]. In rice, the involvement of SA in induced disease resistance has not been clarified because of the constant accumulation of SA in healthy rice leaves [39]. However, Shimono et al. reported that SA application increases OsWRKY45 gene expression in rice [27]. Iwai et al. reported that SA induces resistance against rice blast fungus by exogenous application of SA on top-expanding leaves of 8th leaf-stage “adult” rice [40]. Together, these studies clearly indicate the contribution of SA to IR in rice.
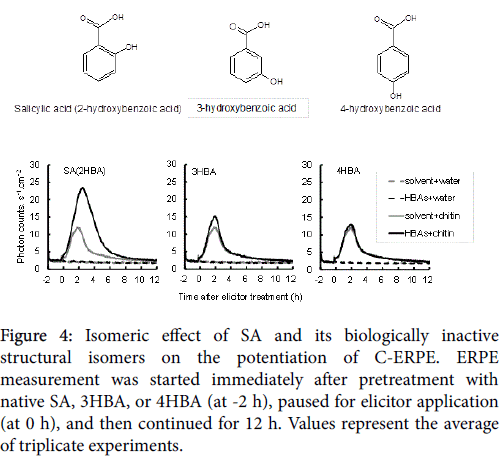
As shown in Figure 3, OsWRKY45 upregulation and potentiation of C-ERPE by SA was observed in WT cells, but it was diminished in the OsWRKY45 -RNAi lines. As shown in Figure 3A (gray columns), the average expression rate compared with WT cells was 55.4% in OsWRKY45 -RNAi line #1. As shown in Figure 3B, the increase in the rate of C-ERPE was 179% in WT cells and 37% in OsWRKY45 -RNAi line #1. As shown in Figure 4, native SA enhanced the C-ERPE (156% increase), whereas the structural isomers of SA (3HBA and 4HBA) had almost no effect on priming (increases of 24% by 3HBA and 10% by 4HBA,). The isomeric effect of SA and its biologically inactive structural isomers has been reported in studies on SAR to show the specificity or structural requirement for SAR induction [41,42], and these effects are confirmed here. These results of experiments to interrupt SA signaling indicate that the enhancement of C-ERPE requires intrinsic hormonal signaling for defense, at least for priming by SAR inducers.
Figure 3: SA-induced potentiation of C-ERPE was diminished by OsWRKY45 knockdown. (A) WT and OsWRKY45 -RNAi cell lines were pretreated with 200 μM SA (gray columns) or solvent (white columns). Two hours after pretreatment, 50 mg aliquots of cells were collected for quantitative RT-PCR of OsWRKY45 and Ubq1 sequences just before chitin elicitor treatment. (B) ERPE measurement was started immediately after pretreatment (at -2 h), paused for elicitor application (at 0 h), and then continued for 12 h. Values represent the average of triplicate experiments. Error bars represent the standard deviation.
Figure 4: Isomeric effect of SA and its biologically inactive structural isomers on the potentiation of C-ERPE. ERPE measurement was started immediately after pretreatment with native SA, 3HBA, or 4HBA (at -2 h), paused for elicitor application (at 0 h), and then continued for 12 h. Values represent the average of triplicate experiments.
Although the authors focused on OsWRKY45 in this study, rice SA signaling branches into OsWRKY45 and NH1, the rice analogue of NPR1 in dicots [43,44]. NPR1 is the central regulator of SAR in dicots, and more than 99% of ASM (BTH)-responsive genes are regulated by NPR1 [45]. However, most ASM (BTH)-responsive upregulated genes are OsWRKY45 dependent, and most ASM-responsive downregulated genes are NH1 dependent in rice [43,44]. The precise role of both regulators in the potentiation of C-ERPE is unclear, although the participation of OsWRKY45 in the potentiation of C-ERPE is irrefutable.
Signal-disruption experiments involving several hormones were next performed to help clarify the contribution of each hormonal cascade to C-ERPE potentiation.
Properties of C-ERPE as a priming detector in rice
The biphasic intensity transition in C-ERPE, in which the first phase is shorter than the second, is a characteristic feature of this system in rice [12]. After chitin perception, phosphatidic acid (PA), a messenger phospholipid, is generated biphasically and induces a burst of reactive oxygen species (ROS) production [46,47]. The authors previously reported that suppression of the phospholipase D (PLD)-mediated second phase of PA generation induced a decrease in the second phase of C-ERPE, whereas exogenous PA application to rice cells mimicked the second phase of C-ERPE [12]. The intensity changes in C-ERPE were proportional to ROS generation, and ROS scavenging lowered the C-ERPE [12-14]. Thus, C-ERPE in rice cells is generated through PA signaling that is closely linked to ROS generation.
Zhan and Xiao [48] proposed biphasic signal amplification of the “PA-ROS-SA” module. In their proposed pathway, pathogen-associated molecular patterns or effector perceptions trigger the first phase of ROS production (0.5-2 h after perception), and this first phase potentiates the following phase (2-10 h after perception). Applications of SA or functional analogues of SA increased C-ERPE and interruptions to SA signaling attenuated this C-ERPE enhancement (See Figures 1B, 2B, and 3B for OsWRKY45 -RNAi; and Figure 4 for application of inactive HBAs). This suggests that amplification driven by the PA-ROS-SA signal cascade may drive the biphasic generation of C-ERPE in rice cells.
On the one hand, C-ERPE potentiation occurs through SA signaling induced by SA or its functional analogues; on the other hand, other molecules such as methyl jasmonate and ethylene can also prime rice cells for C-ERPE amplification [30]. The chitin response in rice is accompanied by an increase in jasmonic acid synthesis [49]. To explain these complexities, the nature of PLDs and PLD-derived PAs that act as versatile signaling components by being incorporated in various kinds of hormonal signaling cascades and by functional overlapping with each other should be considered [50,51]. Further elucidation of the mechanism underlying ERPE potentiation might accelerate the development of multipurpose priming detectors for plant activators, which may be applicable to several biotic and/or abiotic stress disorders, and may help to unlock the complexities of PLD-derived PAs and ROS signaling in plants.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The structure and transformation process of pANDA-OsWRKY45-RNAi.
Supplemental Figure S2. Schema of 5 h-integrated ERPE counts after subtraction.
Supplemental Figure S3. Ten-day-old wild type (left) and OsWRKY45-RNAi cell line (right).
Acknowledgment
This study was supported by a research grant from Shizuoka Prefecture and the Industrial Technology Research Grant Program in FY2007 (grant number: 05A38506a).
References
- Calpe C (2006) Rice international commodity profile. FAO.
- Juliano BO (1993) Rice in human nutrition. FAO.
- Valipour M, Ahmadib MZ, Raeini-Sarjazb M,Sefidkouhib MAG, Shahnazarib A, et al. (2015) Agricultural water management in the world during past half century. Archives Agronomy Soil Sci 61: 657-678.
- Valipour M (2015) Land use policy and agricultural water management of the previous half of century in Africa. Appl Water Sci 5: 367-395.
- Valipour M (2015) Future of agricultural water management in Africa. Archives Agronomy Soil Sci 61: 907-927.
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 11: 793-800.
- Sasaki T, Matsumoto T, Antonio BA, Nagamura Y (2005) From Mapping to Sequencing, Post-sequencing and Beyond. Plant Cell Physiol 46: 3-13.
- Matsumoto T, Wu J, Itoh T, Numa H, Antonio B, et al. (2016) The Nipponbare genome and the next generation of rice genOmics research in Japan. Rice 9: 33.
- Makino T, Kato K, Iyozumi H, Honzawa H, Tachiiri Y, et al. (1996) Ultraweak luminescence generated by sweet potato and Fusarium oxysporum interactions associated with a defense response. Photochem Photobiol 64:953-956.
- Iyozumi H, Kato K, Makino T (2002) Spectral shift of ultraweak photon emission from sweet potato during a defense response. Photochem Photobiol 75: 322-325.
- Iyozumi H, Kato K, Kageyama C, Inagaki H, Yamaguchi A, et al. (2005) Plant defense activators potentiate the generation of elicitor-responsive photon emission in rice. Physiol Mol Plant Pathol 66: 68-74.
- Kageyama C, Kato K, Iyozumi H, Inagaki H, Yamaguchi A, et al. (2006) Photon emissions from rice cells elicited by N-acetylchitooligosaccharide are generated through phospholipid signaling in close association with the production of reactive oxygen species. Plant Physiol Biochem 44: 901-909.
- Kageyama C, Kato K, Inagaki H, Iyozumi H (2007) Participation of hydrogen peroxide in elicitor-responsive photon emission from rice cells. Jpn J Phytopathol 73: 300-303.
- Kato K, Honzawa H, Iyozumi H, Nukui H (2010) Quantitative correlation between elicitor-responsive photon emission and hydrogen peroxide, induced with N-acetylchitohexaose. Jpn J Phytopathol 76: 142-148.
- Kageyama C, Kato K, Iyozumi H, Inagaki H, Yamaguchi A, et al. (2007) The properties of elicitor-responsive photon emissions enhanced by pretreatment with plant defense activators. Jpn J Phytopathol 73: 15-20.
- Bennett M, Mehta M, Grant M (2005) Biophoton imaging: a nondestructive method for assaying R gene responses. Mol Plant Microbe Interact 18: 95-102.
- Kobayashi M, Sasaki K, Enomoto M, Ehara Y (2007) Highly sensitive determination of transient generation of biophotons during hypersensitive response to cucumber mosaic virus in cowpea. J Exp Bot 58:465-472.
- Kessmann H, Stub T, Hofmann C, Maetzke T, Herzog J (1994) Induction of systemic acquired resistance in plants by chemicals. Annu Rev Phytopathol 32: 439-459.
- Tally A, Oostendrop M, Lawton K, Stub T, Bassi B (1999) Commercial development of elicitors of induced resistance to pathogens, in Agrawal AA, Tuzun S, Bent E (eds) Induced plant defenses against pathogens and herbivores. Minnesota APS Press 357-369.
- Narusaka M, Abe H, Kobayashi M, Kubo Y, Kawai K, et al. (2006) A model system to screen for candidate plant activators using an immune-induction system in Arabidopsis. Plant Biotechnol 23: 321-327.
- Ono S, Tanaka T, Watakabe Y, Hiratsuka K (2004) Transient assay system for the analysis of PR-1a gene promoter in tobacco BY-2 cells. Biosci Biotechnol Biochem 68: 803-807.
- Ono S, Kusama M, Ogura R, Hiratsuka (2011) Evaluation of the use of the tobacco PR-1apromoter to monitor defense gene expression by the luciferase bioluminescence reporter system. Biosci Biotechnol Biochem 75: 1796-1800.
- Noutoshi Y, Okazaki M, Kida T, Nishina Y, Morishita Y, et al. (2012) Novel plant immune-priming compounds Identified viahigh-throughput chemical screening target salicylicacid glucosyltransferases in Arabidopsis. Plant Cell 24: 3795-3804.
- Conrath U, Pieterse CMJ, Mauch-Mani B (2002) Priming in plant-pathogen interactions. Trends Plant Sci 7: 210-216.
- Prime-A-Plant Group, Conrath U, Beckers GJ, Flors V, García-Agustín P, et al. (2006) Priming: Getting ready for battle. Mol Plant Microbe Interact 19: 1062-1071.
- Conrath U, Beckers GJM, Langenbach CJG, Jaskiewicz MR (2015) Priming forenhanced defense. Ann Rev Phytopathol 53:97-119.
- Shimono M, Sugano S, Nakayama A, Jiang CJ, Ono K, et al. (2007) Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19:2064-2076.
- Nakayama A, Fukushima S, Goto S, Matsushita A, Shimono M, et al. (2013) Genome-wide identification of WRKY45-regulated genes that mediate benzothiadiazole-induced defense responses in rice. BMC Plant Biol 13:150-161.
- Akagi A, Fukushima S, Okada K, Jiang CJ, Yoshida R, et al. (2014) WRKY45-dependent priming of diterpenoid phytoalexin biosynthesis in rice and the role of cytokinin in triggering the reaction. Plant Mol Biol 86:171-183.
- Kato K, Iyozumi H, Kageyama C, Inagaki H, Yamaguchi A, et al.(2014) Application of ultra-weak photon emission measurements in agriculture. J Photochem Photobiol B: Biol 139: 54-62.
- Kuchitsu K, Kikuyama M, Shibuya N (1993) N-acetylchitooligosaccharides, biotic elicitor for phytoalexin production, induce transient membrane depolarization in suspension-cultured rice cells. Protoplasma 174:79-81.
- Miki D, Shimamoto K (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45: 490-495.
- Miki D, Itoh R, Shimamoto K (2005) RNA silencing of single and multiple members in a gene family of rice. Plant Physiol 138: 1903-1913.
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271-282.
- Görlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, et al. (1996) Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8: 629-643.
- Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, et al. (1996) Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J 10: 71-82.
- Yasuda M, Nakashita H, Yoshida S (2004) Tiadinil, a Novel class of activator of systemic acquired resistance, induces defense gene expression and disease resistance in Tobacco. J Pest Sci 29: 46-49.
- Yasuda M, Kusajima M, Nakajima M, Akutsu K, Kudo T, et al. (2006) Thiadiazole carboxylic acid moiety of tiadinil, SV-03, induces systemic acquired resistance in tobacco without salicylic acid accumulation. J Pest Sci 31: 329-334.
- Yang Y, Qi M, Mei C (2004) Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J 40: 909-919.
- Iwai T, Seo S, Mitsuhara I, Ohashi Y (2007) Probenazole-induced accumulation of salicylic acid confers resistance to Magnaporthe grisea in adult rice plants. Plant Cell Physiol 48: 915-924.
- Conrath U, Chen Z, Ricigliano JR, Klessig DF (1995) Two inducers of plant defense responses, 2,6-dichloroisonicotinec acid and salicylic acid, inhibit catalase activity in tobacco. PNAS 92: 7143-7147.
- Thulke O, Conrath U (1998) Salicylic acid has a dual role in the activation of defense-related genes in parsley. Plant J 14: 35-42.
- Takatsuji H, Jiang CJ, Sugano S (2010) Salicylic Acid Signaling Pathway in Rice and the Potential Applications of Its Regulators. JARQ 44: 217-223.
- Sugano S, Jiang CJ, Miyazawa S, Masumoto C, Yazawa K, et al. (2010) Role of OsNPR1 in rice defense program as revealed by genomewide expression analysis. Plant Mol Biol 74:549-562.
- Wang D, Amornsiripanitch N, Dong X (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog 2:e123.
- Yamaguchi T, Minami E, Shibuya N (2003) Activation of phospholipases by n-acetylchitooligosaccharide elicitor in suspension-cultured rice cells mediates reactive oxygen generation. Physiol Plant 118: 361-370.
- Yamaguchi T, Minami E, Ueki J, Shibuya N (2005) Elicitor-induced activation of phospholipases plays an important role for the induction of defense responses in suspension-cultured rice cells. Plant Cell Physiol 46: 579-587.
- Zhan Q, Xiao S (2015) Lipids in salicylic acid-mediated defense in plants: focusing on the roles of phosphatidic acid and phosphatidil inositol 4-phosphate. Frontiers Plant Sci 6: Article 387.
- Desaki Y, Otomo I, Kobayashi D, Jikumaru Y, Kamiya Y, et al. (2012) Positive crosstalk of MAMP signaling pathways in Rice Cells. PLoS ONE 7: e51953.
- Testerink C, Munnik T (2006) Phosphatidic acid: a multifunctional stress lipid in plants. Trends Plant Sci 10: 368-375.
- Zhao J (2015) Phospholipase D and phosphatidic acid in plant defense response: from protein-protein and lipid-protein interactions to hormone signaling. J Exp Bot 66: 1721-1736.
Relevant Topics
- Basmati Rice
- Drought Tolerence
- Golden Rice
- Leaf Diseases
- Long Grain Rice
- Par Boiled Rice
- Raw Rice
- Rice
- Rice and Aquaculture
- Rice and Nutrition
- Rice Blast
- Rice Bran
- Rice Diseases
- Rice Economics
- Rice Genome
- Rice husk
- Rice production
- Rice research
- Rice Yield
- Sticky Rice
- Stress Resistant Rice
- Unpolished Rice
- White Rice
Recommended Journals
Article Tools
Article Usage
- Total views: 11454
- [From(publication date):
December-2016 - Jul 18, 2025] - Breakdown by view type
- HTML page views : 10539
- PDF downloads : 915