Characterization of Influenza Viruses Responsible for Acute Respiratory Illness in Cambodia from 2015 to 2016
Received: 11-Dec-2019 / Accepted Date: 24-Dec-2019 / Published Date: 31-Dec-2019 DOI: 10.4172/2332-0877.1000413
Abstract
Background: Acute Respiratory Infection (ARI) is a leading cause of morbidity and mortality worldwide. During a study describing the circulation and the prevalence of respiratory viruses and bacteria in Cambodia, an ancillary analysis focussed on influenza viruses.
Method: Between July 2015 and December 2016, 18 to 50-year-old patients presenting with ARI (cough+sore throat+fever ≥ 37.5°C) and attending four referral hospitals were included. In each site, out-patients, in-patients and non-ARI controls (outpatients with non-infectious diseases) were recruited. Clinical information and nasal washes were collected. Influenza and other respiratory pathogens were screened by multiplex real-time PCR. Each influenza virus detected was subsequently typed, subtyped, cultured, tested for Neuraminidase Inhibitors susceptibility and sequenced by direct whole genome sequencing.
Results: Amongst the 1006 included patients, 48 were positive for influenza (5.4%), including 36 Influenza A (19 A(H1N1)pdm09 and 17 A(H3N2)), 11 Influenza B (9 Yamagata and 2 Victoria) and one A(H1N1)pdm09+B Victoria co-infection. Overall, 11 in-patients (6 H1N1pdm09, 2 H3N2, 2 B and 1 H1N1pdm09+B co-infection), 36 out-patients (12 A(H1N1)pdm09, 15 A(H3N2) and 9 B) and 1 control (1 H1N1pdm09) were positive. These viruses circulated year-round with 2 peaks during the rainy season (August 2015 and June 2016), and a switch from A(H3N2) to BYamagata
and to A(H1N1)pdm09 with almost no overlap was observed. All viruses were similar to the vaccine strains, and susceptible to NAI.
Conclusion: We report a low prevalence of influenza in this adult population (5.4%). Most cases were due to
influenza A (77.1%) with a balanced distribution between A(H1N1)pdm09 and A(H3N2) (20 vs. 17), and a higher
proportion of A(H1N1)pdm09 in in-patients. No molecular difference was observed between viruses of in- and outpatients,
and no resistance was detected. The year-round circulation and virus switch is similar to this reported in
other sub-tropical areas.
Keywords: Influenza viruses; Acute respiratory infection; Adults;Cambodia
Introduction
Acute Respiratory Infection (ARI) is a leading cause of morbidity and mortality worldwide. Influenza viruses are a frequent aetiology for ARI [1,2]. Each year, an estimated 5%–10% of adults and 20%– 30% of children are infected with influenza, resulting in 3–5 million cases of severe disease and approximately 1 million deaths worldwide [3]. Despite the availability of vaccines and specific therapeutic agents, the influenza burden remains very high worldwide [4,5].
Southeast Asia is a region of interest for influenza epidemiology and ecology, with a high burden of disease, complex transmission patterns, and a potential risk for infection with avian viruses [6]. Since 2003 and the emergence of A(H5N1) in the wild birds as well as in the domestic poultry, the zoonotic risk for transmission of avian influenza viruses to humans has been scrutinized, and enhanced surveillance and mitigation measures have been implemented in this region to reduce the risk for cross-barrier transmission of an avian influenza virus. In the context of endemic zoonotic influenza, robust influenza surveillance in this region is important to detect rapidly human cases with avian virus, and to assess the risk for pandemic, either through direct adaptation [7] of via genetic reassortment [8,9].
For some years now, conventional culture methods and recent molecular techniques have been implemented and subsequently used for accurate diagnosis and surveillance of influenza in south-east Asia [10]. However, there is a remaining need for the dissemination of more accessible diagnostic tools for influenza surveillance, disease management, reduce unnecessary lab testing or treatments and allow rapid implementation of NAI treatment in severe cases [11]. Molecular testing has proved to be cost-effective for all these purposes [12,13].
The proactive surveillance for seasonal influenza can provide pattern of virus circulation and characteristics of seasonal epidemics based on comprehensive surveillance in the general population. It helps for the description of influenza circulation and burden at a national and regional level, it provides insights into the genetic and phenotypic evolution of influenza viruses and allows optimization of prevention and control strategies [13-15].
Here, we describe the circulation of influenza in the 18-50 agegroups during an 18 month period between 2016-2017. The viruses detected from in- and out-patients were characterized through conventional NIC techniques (HAI, RT-PCR), analysed for their susceptibility to antivirals (NAI), and compared through direct full genome sequencing to viruses reported at the same period of time in South-East Asia.
Methods
Study design
The acute respiratory illness (ARI) study was conducted in four hospitals: (1) Serei Saophoan Referral Hospital at Banteay Mean Chey at north-west 359 Km from Phnom Penh, (2) Kampong Chhnang Provincial Hospital at Kampong Chhnang province situate at northwest 91 km from Phnom Penh, (3) Bati Referral Hospital at Takeo Province situate 78 Km from south of Phnom Penh and Kep Referral Hospital at Kep province situate 174 Km from south of Phnom Penh as described in (Figure 1). In- and out-patients aged between 18 and 50 presenting with ARI symptoms (cough/sore throat) plus fever (≥ 37.5°C) were recruited. Patients visiting the same hospitals as outpatient for non-ARI illness were also recruited as controls.
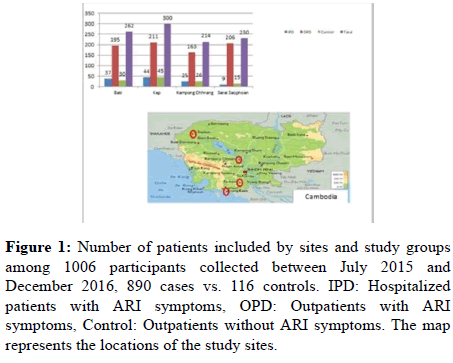
Figure 1: Number of patients included by sites and study groups among 1006 participants collected between July 2015 and December 2016, 890 cases vs. 116 controls. IPD: Hospitalized patients with ARI symptoms, OPD: Outpatients with ARI symptoms, Control: Outpatients without ARI symptoms. The map represents the locations of the study sites.
Clinical data collection, sample collection and shipment
For each patient, the objectives of the study were explained and an informed consent was obtained. Clinicians filled out a study case report form (CRF) for each participant including demographic and clinical data (age, gender, residence, occupation, onset of symptoms, date and site of submission, clinical diagnosis, smoking, illness information, history of contacts with animals, antibiotic treatment), and a nasal wash was collected by a trained healthcare worker. Samples were collected in universal transport medium (UTM) and stored at +4°C until shipment to the University of Health Sciences (UHS) laboratory based in Phnom Penh. Upon arrival, all specimens were checked for sample integrity and subsequently aliquoted and stored at -80°C for further analysis.
Ethical Statement
This study protocol was approved by the National Ethic Committee Health Research of Cambodia (Ref: 219NECHR date 29 June 2015). Samples were collected after an informed written consent obtained from each patient.
Molecular assays
Total nucleic acids were extracted from the specimen using Exgene ™ Viral DNA/RNA kit (AITbiotech, Singapore). Influenza virus and other respiratory pathogens were detected by multiplex real-time PCR (abTESTM ARIS panel, AITbiotech, Singapore) on Bio-Rad CFX96, according to the manufacturer. The positive samples for influenza virus were subsequently typed and subtyped using abTES ™ Flu4 qPCR I kit (AITbiotech, Singapore) on Bio-Rad CFX96.
Influenza virus isolation and antigenic characterization
All of the specimens tested positive for Influenza by PCR were sent to the Centre National de Référence des Virus des Infections Respiratoires, Lyon for further analysis. Each specimen was inoculated on MDCK cells in the presence of trypsin (SIGMA Life Science, Darmstadt, Germany). The isolates were characterized by a Hemagglutination Inhibition Assay (HIA) using reference antigens and anti-sera provided either by the WHO cc (Crick Institute, London, UK) or prepared on site (NIC, Lyon).
Neuraminidase inhibitors susceptibility assays
Oseltamivir carboxylate was provided by Hoffmann-La Roche (Roche Diagnostics GmbH, Mannheim, Germany) and zanamivir by GlaxoSmithKline (GSK, Brentford, UK). The fluorometric inhibition assays were performed using a MFX fluorometer (Dynex technologies, Chantilly, VA, USA) as previously described [16]. The concentrations of drug required to inhibit 50% of the NA activity (IC50) were calculated using Sigma Plot software v 8.0 (Systat software, SanJose, CA, USA).
RNA extraction, viral load determination, and full-genome amplification
RNA was directly extracted from primary specimen using an EMAG® automated extraction platform (bioMérieux, Marcyl ’ Etoile, France). In order to lower human and bacterial DNA content of the extract, nucleic acids were incubated with 1 μL of Turbo DNAse, 2 μL of Turbo DNAse buffer (Life Technologies, Carlsbad, CA, USA), and 0.5 μL of RNasin plus RNase Inhibitor (Promega Corporation, Madison, WI, USA) for 90 min at 37°C before stopping the enzymatic reaction through immediate purification by magnetic beads (0.5x; NucleoMag® NGS Clean-up and Size Select, Macherey – Nagel, Düren, Germany). Each segment was retrotranscribed and amplified using a multi-segment reverse transcription protocol, optimized by adding 0.5 μl of RNasin plus RNase Inhibitor [17].
Phylogenetic analysis
Evolutionary analyses of HA and NA gene sequences were carried out with all Cambodian influenza viruses and other related viruses collected worldwide. Reference strain sequences as well as sequences from viruses collected within the study period in various other South- East Asian countries (Vietnam, Laos, Thailand, Myanmar and Philippines) were obtained from EpiFlu Database available via the Global Initiative on Sharing all Influenza Data (GISAID) website (www.gisaid.org) and from NCBI Influenza Resource (www.ncbi.nlm.nih.gov/genomes/flu/) and included in the analysis. Sequences were aligned using MUSCLE program implemented in Seaview version 4. Best models were determined from alignments in www.iqtree.org. Maximum likelihood (ML) phylogenetic trees were generated using PhyML as implemented in Seaview version 4. Approximate likelihood ratio test (aLRT) values >95% were shown on the branch. ML phylogenies were annotated using FigTree version 1.4.3. Aligned sequences were exported to Molecular Evolutionary Genetics Analysis (MEGA) version 7.0.26 in order to determine the mutations responsible for changes in the topologies.
Results
Demographic and clinical characteristic of participants at enrolment
Between July 2015 and December 2016, a total of 1006 participants were prospectively enrolled in the surveillance study, in which 890 patients presented ARI symptoms, including 775 out-patients, 115 hospitalized in-patients, and 116 healthy controls. Of the total study population, 262 (26.04%), 300 (29.82%), 214 (21.27%) and 230 (22.86%) were enrolled in Bati referral hospital, Kep referral hospital, Kampong Chhnang referral hospital and Serei Saophoan referral hospital, respectively (Figure 1).
Pathogen identified by molecular-based sub-typing
For influenza virus detection, 48/890 patients (5.4%) were positive, including 36 Influenza A, 11 Influenza B and 1 Influenza A and B coinfection. Of the 36 Influenza A positive, 19 had H1N1pdm09 and 17 H3N2. The 11 Influenza B positive were 9 Yamagata lineage and 2 Victoria lineage viruses. Overall, 11/48 were in-patients (2 A(H3N2) (18.2%), 6 A(H1N1)pdm09 (54.5%) and 2 B Yamagata (18.2%)), while 37/48 were out-patients (15 A(H3N2) (40.5%), 13 A(H1N1)pdm09 (35.1%), 7 B Yamagata (18.9%) and 2 B Victoria (5.4%)). The single co-infection observed was a A(H1N1)pdm09 and B-Victoria co-infection from an in-patient. In the control group, only one influenza A(H1N1)pdm09 virus was detected.
Influenza virus circulation
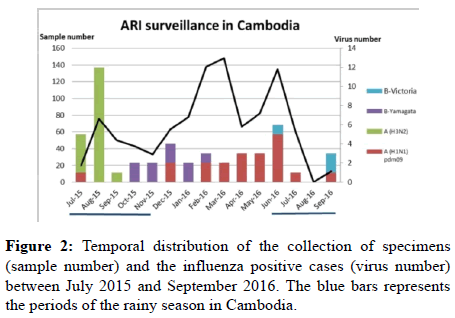
The influenza viruses circulated sequentially, starting with A (H3N2) between July to September 2015, followed by the B-Yam lineage (Oct 15 to Feb 16) and then A(H1N)pdm09 (Dec 15 to Sept 16). Influenza viruses have been detected throughout the 15 months of the study (Figure 2).
Influenza virus isolation and antigenic characterization
Among the 48 positive samples, 44 viruses were isolated on MDCK cell culture and characterized by HIA, including 17 A(H1N1)pdm09, 15 A(H3N2), 9 B-Yam and 3 B-Vic strains. A (H1N1)pdm09 viruses were either related to A/California/7/2009 (1/17) or to A/Michigan/ 45/2015 (16/17) strains. All 15 A (H3N2) strains were undistinguishable from A/Hong Kong/4801/2014, all 3 B-Vic viruses were identical to B/Brisbane/60/2008, and amongst the 9 B-Yam viruses, 8 were identical to B/Massachusetts/2/2012 and 1 to B/ Phuket/3073/2013.
Neuraminidase Inhibition (NI) assay result
All 44 isolated viruses were tested for susceptibility to oseltamivir and zanamivir. The IC50 values of the 17 A (H1N1) pdm09 were between 0.15 – 0.55 nM for oseltamivir and between 0.04-1.35 nM for zanamivir. The fifteen Influenza A (H3N2) had IC50 values between 0.12–1.42 nM for oseltamivir and between 0.49-2.40 nM for zanamivir. The IC50 values of the 12 Influenza B were between 5.45-14.10 nM for oseltamivir and between 1.25-3.43 nM for zanamivir. All the tested strains were susceptible to oseltamivir.
Phylogenetic analysis of the HA and NA genes
HA and NA genes were directly sequenced from the clinical specimen, to avoid irrelevant substitutions related to MDCK passages. Overall, 10 A(H1N1)pdm09, 12 A(H3N2), 3 B-Vic and 9 B-Yam could be sequenced and analyzed. The designation of the clades for HA and NA phylogenetic trees of influenza viruses is based on WHO reports (Crick Institute, WHO Collaborative Centre).
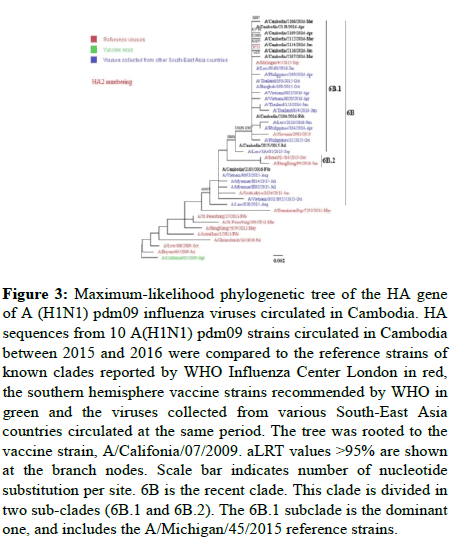
A(H1N1)pdm09: As shown in Figure 3, all A (H1N1) pdm09 viruses fell into genetic group 6B, mainly in subgroup 6B.1 (except for A/Cambodia/2105/2016). As compared to the A/California/ 07/2009, A (H1N1)pdm09 reference strain, the Cambodian strains displayed the 6B clade-specific amino acid substitutions (P83S, D97N, S185T, S203T, K163Q, A256T, K283E, I321V) in HA1 and in HA2 (E47K, S124N, E172K), along with S84N, S162N, I216T 6B.1 specific substitutions. Overall, 6 deduced amino acid changes (V47I, S69T, K171R, A187T and E235D in HA1 and N71S in HA2) were found in the HA of the Cambodian A(H1N1)pdm09 viruses. The phylogenetic tree of NA genes showed a close proximity with A/Michigan/45/2015 except for A/Cambodia/2105/2016 (data not shown).
Figure 3: Maximum-likelihood phylogenetic tree of the HA gene of A (H1N1) pdm09 influenza viruses circulated in Cambodia. HA sequences from 10 A(H1N1) pdm09 strains circulated in Cambodia between 2015 and 2016 were compared to the reference strains of known clades reported by WHO Influenza Center London in red, the southern hemisphere vaccine strains recommended by WHO in green and the viruses collected from various South-East Asia countries circulated at the same period. The tree was rooted to the vaccine strain, A/Califonia/07/2009. aLRT values >95% are shown at the branch nodes. Scale bar indicates number of nucleotide substitution per site. 6B is the recent clade. This clade is divided in two sub-clades (6B.1 and 6B.2). The 6B.1 subclade is the dominant one, and includes the A/Michigan/45/2015 reference strains.
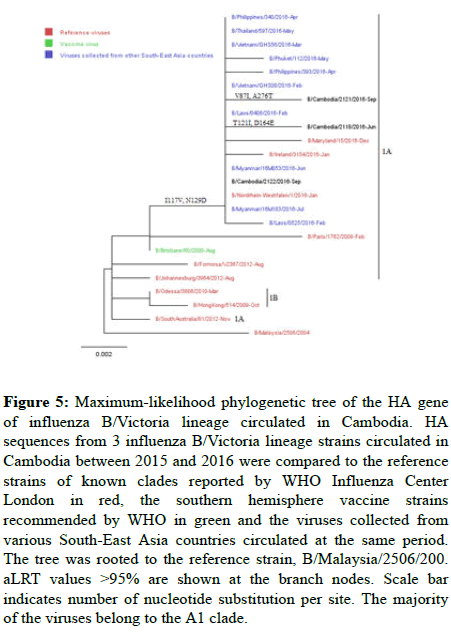
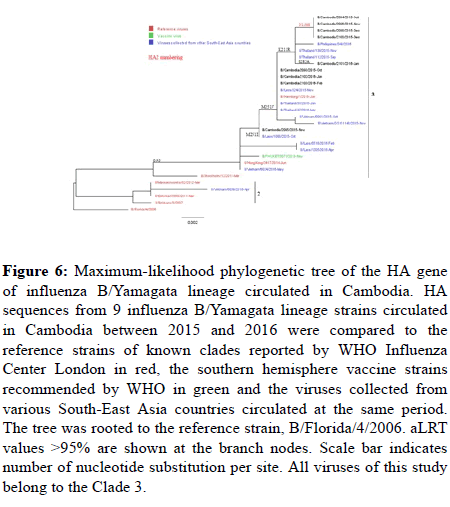
Influenza B: All B-Vic Cambodian viruses belonged to clade 1A, as the B/Brisbane/60/2008 reference strain (Figure 4 and 5), and all 9 B/ Yamagata lineage viruses belonged to clade 3, as the B/Phuket/ 3073/2013 reference strain (Figure 6).
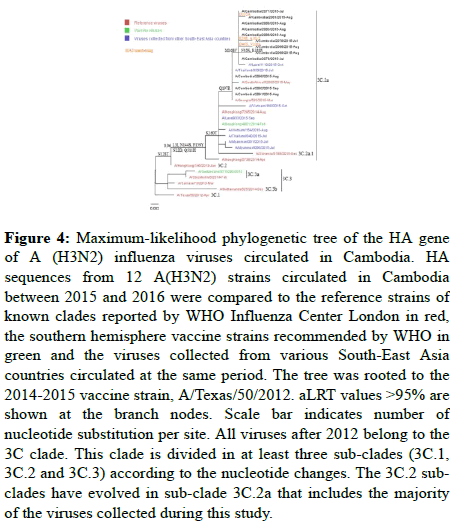
Figure 4: Maximum-likelihood phylogenetic tree of the HA gene of A (H3N2) influenza viruses circulated in Cambodia. HA sequences from 12 A(H3N2) strains circulated in Cambodia between 2015 and 2016 were compared to the reference strains of known clades reported by WHO Influenza Center London in red, the southern hemisphere vaccine strains recommended by WHO in green and the viruses collected from various South-East Asia countries circulated at the same period. The tree was rooted to the 2014-2015 vaccine strain, A/Texas/50/2012. aLRT values >95% are shown at the branch nodes. Scale bar indicates number of nucleotide substitution per site. All viruses after 2012 belong to the 3C clade. This clade is divided in at least three sub-clades (3C.1, 3C.2 and 3C.3) according to the nucleotide changes. The 3C.2 subclades have evolved in sub-clade 3C.2a that includes the majority of the viruses collected during this study.
Figure 5: Maximum-likelihood phylogenetic tree of the HA gene of influenza B/Victoria lineage circulated in Cambodia. HA sequences from 3 influenza B/Victoria lineage strains circulated in Cambodia between 2015 and 2016 were compared to the reference strains of known clades reported by WHO Influenza Center London in red, the southern hemisphere vaccine strains recommended by WHO in green and the viruses collected from various South-East Asia countries circulated at the same period. The tree was rooted to the reference strain, B/Malaysia/2506/200. aLRT values >95% are shown at the branch nodes. Scale bar indicates number of nucleotide substitution per site. The majority of the viruses belong to the A1 clade.
Figure 6: Maximum-likelihood phylogenetic tree of the HA gene of influenza B/Yamagata lineage circulated in Cambodia. HA sequences from 9 influenza B/Yamagata lineage strains circulated in Cambodia between 2015 and 2016 were compared to the reference strains of known clades reported by WHO Influenza Center London in red, the southern hemisphere vaccine strains recommended by WHO in green and the viruses collected from various South-East Asia countries circulated at the same period. The tree was rooted to the reference strain, B/Florida/4/2006. aLRT values >95% are shown at the branch nodes. Scale bar indicates number of nucleotide substitution per site. All viruses of this study belong to the Clade 3.
Discussion
During the study, influenza viruses were detected in all sites, all through the network, with a year round circulation as previously described [18,19]. As expected, most cases were detected during the rainy season with a peak in August 2015 and in June 2016 corresponding at the beginning of the rainy season (Figure 2). The reported prevalence of influenza in the study was low (5.4%). This may be due to the age-group analyzed in this study (18-50 years of age) where the influenza incidence is usually reported to be low, as compared to children [2,20].
Most of Influenza cases were due to Influenza A viruses (77.1%), with an almost equal distribution between A(H1N1)pdm09 and A(H3N2) (20 (54%) vs. 17 (46%), respectively). The A(H3N2) viruses circulated in 2015, from July to September, then were displaced first by the B viruses that emerged from October till February 2016, and eventually by A(H1N1)pdm09 at the end of the study between December 2015 and September 2016. This sequential circulation of the virus types/subtypes (influenza waves) has already been described in previous studies conducted in the neighboring areas of Western Cambodia [2].
In our study, A(H3N2) was more frequently detected in the outpatient population (40.5% vs. 18.2%), whereas A(H1N1)pdm09 was more frequent in in-patients (54.5% vs. 35.1%). This predominance of more severe cases (i.e. requiring hospitalization) related to A(H1N1)pdm09 in the 18-50 age-group has also been reported in Europe and America [21,22].
The incidence of the B viruses was very low. This was not surprising as the majority of influenza B virus infection is reported in children and adolescent between 5 – 19 years [23]. However, as reported in Thailand, the influenza B-Yam strains were more frequent in 2015 than in 2016, and the B-Vic strains were detected in 2016 at the end of the surveillance [24].
During this study, the influenza virus detection by RT-PCR was carried out in Phnom Penh, in the Rodolphe Merieux lab, and the isolation and sequencing were obtained in the NIC in Lyon. Among the 48 positive samples by real-time RT-PCR, 44 have been isolated on MDCK cell culture. All these viruses were subsequently characterized by HAI, and 34 could be processed for HA and NA genetic analysis by NGS. There was no evidence of introduction of avian viruses in the patients included in the study.
Phylogenetic analysis of the HA gene showed that all influenza A(H3N2) strains in Cambodia belonged to genetic clade 3C.2a, similar to the A/Hong Kong/4801/2014 reference strain which was the 2016 vaccine strain for southern hemisphere (Figure 4). The phylogenetic analysis showed viruses similar to these reported in Thailand in 2015 [18]. Similarly, the A(H1N1)pdm09 viruses detected were similar to those detected in the neighboring countries, confirming the circulation of similar strains in this large region of south East Asia [2,14].
Last, all viruses detected and tested for Neuraminidase Inhibitor (NAI) susceptibility did not show any reduced or highly-reduced susceptibility to Oseltamivir. The limited number of viruses tested (44) makes difficult to ascertain for the absence of circulation of any resistant virus, as reported in Thailand Australia and Singapore, where A(H1N1)pdm09 viruses with reduced susceptibility related to the S247N substitution in NA have been described between 2009 and 2010 [25,26] Again, Influenza B viruses with reduced susceptibility related to a D197N substitution have been described in Cambodia in the past [27], but not in our study.
Conclusion
The collection, handling and processing of the clinical specimens in Cambodia confirmed the capacity building and the transfer of technology from the Lyon NIC to the Rodolphe Merieux laboratory in Phnom Penh, Cambodia. Virus isolation remains to be implemented to achieve a complete transfer of technology. The data collected from these 2015-2016 specimens provided detailed information on the circulating viruses, including about the genetic content of these viruses, the comparison of viruses from hospitalized and nonhospitalized patient, their antigenic and genetic characterization, and their susceptibility to antiviral drugs (NAI). This additional capacity will strengthen the influenza surveillance capacity in Cambodia, and will be of help to provide a better picture about the circulation of seasonal influenza in this country, and will be useful for the rapid detection and characterization of sporadic human cases with zoonotic viruses with a pandemic potential.
Authors’ Contributions
BK and MC designed the study, BK and MV did the Biological investigations, BK, MV and BL analysed the results, BK and BL drafted the manuscript, and BK, MV MC and BL corrected the manuscript and prepared the final version.
Acknowledgments
We thank all the patients for their participation, as well as clinicians at study side for their inclusion participant and data collection. We thank the staff of the CNR in Lyon (Gwendolyne Burfin, Clio Soucratous, Nomine Omoni and Remy Fanget) for their technical support.
This study was supported by a combined Grant from the Fondation Mérieux and from the Institut de Recherche et Développement (IRD), and was carried out in collaboration with the CNR des virus des infections respiratoires (dont la grippe) with the financial support of Santé Publique France.
References
- Takahashi K, Suzuki M, Minh le N, Anh N H , Huong LT, et al. (2013) The incidence and aetiology of hospitalised community-acquired pneumonia among Vietnamese adults: A prospective surveillance in Central Vietnam. BMC Infect Dis 13: 296.
- Timmermans A, Melendrez MC, Se Y, Chuang I, Samon N, et al. (2016) Human sentinel surveillance of influenza and other respiratory viral pathogens in border areas of Western Cambodia. PLoS One 11: e0152529.
- Vaccines against influenza WHO position paper-November (2012) Wkly Epidemiol Rec 87: 461-476.
- Heneghan CJ, Onakpoya I, Thompson M, Spencer EA, Jones M, et al. (2014) Zanamivir for influenza in adults and children: Systematic review of clinical study reports and summary of regulatory comments. BMJ 348: g2547.
- Jefferson T, Jones M, Doshi P, Spencer E A, Onakpoya I, et al. (2014) Oseltamivir for influenza in adults and children: systematic review of clinical study reports and summary of regulatory comments. BMJ 348: g2545.
- Anderson K B, Simasathien S, Watanaveeradej V, Weg A L, Ellison D W, et al. (2018) Clinical and laboratory predictors of influenza infection among individuals with influenza-like illness presenting to an urban Thai hospital over a five-year period. PLoS One 13: e0193050.
- De Vries R P, Zhu X, McBride R, Rigter A, Hanson A, et al. (2014) Hemagglutinin receptor specificity and structural analyses of respiratory droplet-transmissible H5N1 viruses. J Virol 88: 768-773.
- Lindh E, Ek-Kommonen C, Vaananen V M, Vaheri A, Vapalahti O, et al. (2014) Molecular epidemiology of H9N2 influenza viruses in Northern Europe. Vet Microbiol 172: 548-554.
- Pascua P N, Choi YK (2014) Zoonotic infections with avian influenza A viruses and vaccine preparedness: A game of "mix and match". Clin Exp Vaccine Res 3:140-148.
- Kim D K, Poudel B, (2013) Tools to detect influenza virus. Yonsei Med J 54: 560-566.
- Lacroix S, Vrignaud B, Avril E, Moreau-Klein A, Coste M, et al. (2015) Impact of rapid influenza diagnostic test on physician estimation of viral infection probability in paediatric emergency department during epidemic period. J Clin Virol 72:141-145.
- Behzadi M A, Ziyaeyan M, Alborzi A, (2016) A diagnostic one-step real-time reverse transcription polymerase chain reaction method for accurate detection of influenza virus type A. Arch Med Sci 12:1286-1292.
- Vemula S V, Zhao J, Liu J, Wang X, Biswas S, Hewlett I (2016) Current Approaches for Diagnosis of Influenza Virus Infections in Humans. Viruses 8: 96.
- Tewawong N, Prachayangprecha S, Vichiwattana P, Korkong S, Klinfueng S, et al. (2015) Assessing Antigenic Drift of Seasonal Influenza A(H3N2) and A(H1N1)pdm09 Viruses. PLoS One 10: e0139958.
- Mosnier A, Caini S, Daviaud I, Bensoussan J L, Stoll-Keller F, et al. (2015) Ten influenza seasons in France: Distribution and timing of influenza A and B circulation, 2003-2013. BMC Infect Dis 15: 357.
- Ferraris O, Kessler N, Lina B, (2005) Sensitivity of influenza viruses to zanamivir and oseltamivir: A study performed on viruses circulating in France prior to the introduction of neuraminidase inhibitors in clinical practice. Antiviral Res 68: 43-48.
- Zhou B, Donnelly M E, Scholes D T, St George K, Hatta M, et al. (2009) Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J Virol 83:10309-10313.
- Tewawong N, Suntronwong N, Vichiwattana P, Vongpunsawad S, Theamboonlers A, Poovorawan Y (2016) Genetic and antigenic characterization of hemagglutinin of influenza A/H3N2 virus from the 2015 season in Thailand. Virus Genes 52:711-715.
- Tewawong N, Suntronwong N, Korkong S, Theamboonlers A, Vongpunsawad S, et al. (2017) Evidence for influenza B virus lineage shifts and reassortants circulating in Thailand in 2014-2016. Infect Genet Evol 47:35-40.
- Hasan R, Rhodes J, Thamthitiwat S, Olsen S J, Prapasiri P, et al. (2014) Incidence and etiology of acute lower respiratory tract infections in hospitalized children younger than 5 years in rural Thailand. Pediatr Infect Dis J 33: e45-52.
- Chakhunashvili G, Wagner A L, Power L E, Janusz C B, Machablishvili, et al. (2018) Severe Acute Respiratory Infection (SARI) sentinel surveillance in the country of Georgia, 2015-2017. PLoS One 13: e0201497.
- Torner N, Martinez A, Basile L, Mosquera M, Anton A, et al. (2018) Descriptive study of severe hospitalized cases of laboratory-confirmed influenza during five epidemic seasons (2010-2015). BMC Res Notes 11: 244.
- Heikkinen T, Ikonen N, Ziegler T (2014) Impact of influenza B lineage-level mismatch between trivalent seasonal influenza vaccines and circulating viruses, 1999-2012. Clin Infect Dis 59:1519-1524
- Tewawong N, Suwannakarn K, Prachayangprecha S, Korkong S, Vichiwattana P, et al. (2015) Molecular epidemiology and phylogenetic analyses of influenza B virus in Thailand during 2010 to 2014. PLoS One 10: e0116302.
- Hurt A C, Lee R T, Leang S K, Cui L, Deng Y M, et al. (2011) Increased detection in Australia and Singapore of a novel influenza A(H1N1)2009 variant with reduced oseltamivir and zanamivir sensitivity due to a S247N neuraminidase mutation. Euro Surveill 16.
- Tewawong N, Vichiwattana P, Korkong S, Klinfueng S, Suntronwong N, et al. (2017) Evolution of the neuraminidase gene of seasonal influenza A and B viruses in Thailand between 2010 and 2015. PLoS One 12: e0175655
- Hatakeyama S, Sugaya N, Ito M, Yamazaki M, Ichikawa, et al. (2007) Emergence of influenza B viruses with reduced sensitivity to neuraminidase inhibitors. Jama 297:1435-1442.
Citation: Ka B, Valette B, Chou M, Lina B (2019) Characterization of Influenza Viruses Responsible for Acute Respiratory Illness in Cambodia from 2015 to 2016. J Infect Dis Ther 7: 413. DOI: 10.4172/2332-0877.1000413
Copyright: © 2019 Ka B, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 2699
- [From(publication date): 0-2019 - Apr 04, 2025]
- Breakdown by view type
- HTML page views: 1928
- PDF downloads: 771