Characterization of an Extracellular Polyhydroxyalkanoate Depolymerase from Streptomyces sp. SFB5A
Received: 31-Aug-2018 / Accepted Date: 11-Sep-2018 / Published Date: 14-Sep-2018 DOI: 10.4172/2155-6199.1000452
Keywords: PHB; PHV; Polyhydroxyalkanoates; PHA depolymerase; Streptomyces ; Catalytic type 1 domain; FnIII linker domain; Substratebinding domain
Introduction
Petroleum-based plastics present two major problems: they require a non-renewable fossil fuel for their manufacture and are recalcitrant to degradation upon disposal. Poly(3-hydroxyalkanoates) (PHAs) are water-insoluble polymers of R-3-hydroxyalkanoic acid monomers in chains of 100 to 3000 produced by many bacteria for storage of carbon and energy [1]. They represent a biodegradable alternative to petroleum-based plastics for certain applications and have been produced commercially on a limited scale. Short-chain-length PHAs (PHASCL) consist of monomers with 3 to 5 carbon atoms [2]. The most common of these is poly(R-3-hydroxybutyrate) (PHB), composed of R-3-hydroxybutanoic acid (3HB) monomers [1]. Another common PHASCL is poly(R-3-hydroxyvalerate) (PHV), composed of R-3- hydroxyvaleric acid (3HV) monomers. Some bacteria produce heteropolymers (PHBV) composed of 3HB and 3HV monomers in various proportions [1]. Medium-chain-length PHAs (PHAMCL), such as poly(R-3-hydroxyoctanoate) (PHO), consist of monomers containing 6 or more carbon atoms [2]. Native, intracellular PHAs (nPHAs) from PHA-producing bacteria have an amorphous structure, which is usually altered upon removal from the cell or chemical treatments, forming denatured, i.e., partially crystalline PHAs (dPHAs) [2]. Amorphous analogs of nPHAs, abbreviated aPHAs, can be prepared by treatment of chloroform solutions of dPHAs with detergents [3].
Many microorganisms degrade dPHAs, using extracellular PHA depolymerases. These are secreted into the surrounding medium and hydrolyze PHAs to their monomers, which are transported into the cells and catabolized. Numerous PHA depolymerases have been purified and characterized (reviewed in references 2 and 4). Biosynthesis of PHA depolymerases occurs during growth on PHAs or their monomers, but is repressed by sugars, organic acids, and amino acids [2,5-8].
Streptomyces is a very broad genus of Gram positive, filamentous bacteria with high % G+C (~70%) that produce abundant reproductive spores and various antibiotics [9]. They degrade a variety of polymers including: agar, starch, cellulose, chitin, and xylan, with degradation catalyzed by polymer-specific hydrolases [10]. In situ soil studies show that numerous Streptomyces species degrade PHB and PHBV as well [11]. Extracellular dPHA depolymerases are reported in several Streptomyces species, some of which specifically degrade PHASCL [7,12-15], while others degrade PHAMCL [16-18].
Despite the taxonomic breadth of Streptomyces , comparatively few extracellular PHA depolymerases from this important genus have been described to date. We report here the isolation of a PHB-degrading actinomycete, Streptomyces sp. SFB5A, the purification and characterization of an extracellular dPHASCL depolymerase from the organism, and the cloning of its associated gene. We show that the enzyme produces mostly monomers with some dimers and trimers during PHA degradation and propose a possible model to explain this result, based on biochemical and structural data.
Materials and Methods
Chemicals
PHB and the sodium salts of R,S-3-hydroxybutanoic acid (R,S-3HB) and R- 3- hydroxybutanoic acid (R-3HB) were obtained from Sigma- Aldrich, St. Louis, MO, USA. PHV was prepared by culturing Chromobacterium violaceum with sodium valerate as described [19]. A PHBV copolymer containing 17 mol% 3HV (trade name Biopol® BX P029) was obtained from Zeneca, London, UK. PHO was prepared by culturing Pseudomonas oleovorans with sodium octanoate as described [20]. Homogeneous suspensions of granular PHB and PHBV (3% to 4% w/v) were prepared by sonication, autoclaved, and stored at 4°C. Sonicated PHV suspensions readily aggregated, requiring considerable care in pipetting. For preparation of PHA thin films, PHAs were made to 1% w/v in chloroform and heated at 50°C until dissolved. Amorphous PHB (aPHB) was prepared as described, with 50 mM sodium cholate as the detergent [3]. All other chemicals were of reagent grade or better.
Culture media and growth conditions
A defined mineral salts medium (SNC) [21] was used for experiments on PHA depolymerase synthesis. Carbon sources were added separately as filter-sterilized or autoclaved stocks. SNC containing 0.2% w/v PHB was referred to as SNC-PHB. Nutrient broth (NB), nutrient agar (NA), tryptic soy broth (TSB), trypticase soy agar (TSA) (Becton Dickinson and Company, Franklin Park, NJ, USA) and mannitol soy flour medium (MSF) [9] were used as complex media. Streptomyces sp. SFB5A was maintained on SNC-PHB agar plates. Spore suspensions were prepared from SNC plus 0.5% glucose agar cultures as described [9]. All agar plate cultures of Streptomyces sp. SFB5A were incubated at 30°C. All broth cultures were incubated with rotary shaking (200 rpm) at 30°C for times indicated.
Isolation of strain SFB5A
Well decayed hardwood mulch (1.0 g) was added to 99.0 mL of sterile 0.85% saline solution in a dilution bottle. The mixture was shaken vigorously for 30 sec and allowed to settle for 1 min. Samples (0.1 mL) of the supernatant liquid were serially diluted and plated onto SNC agar plates previously overlaid with 5 mL of SNC-PHB agar. The plates were incubated at 30°C for 48 h. Selected colonies exhibiting large clearing zones in the opaque PHB overlay were streaked first for isolation on fresh SNC-PHB overlay plates, and then twice on NA plates. One isolate, SFB5A, was chosen for further study. It has been deposited in the Agricultural Research Service (ARS) Open Culture Collection, Peoria, IL, USA as Streptomyces sp. SFB5A, NRRL B-65520.
Taxonomic characterization of strain SFB5A
Genomic DNA of strain SFB 5A was isolated by a salting out method, as described [22]. A portion of the 16S-rRNA gene was amplified from the genomic DNA by the polymerase chain reaction (PCR) with primer pairs StrepB/StrepF [23] and pA/pH [24](Table S1).Reaction mixtures included 100 ng of genomic DNA, 0.5 μM of each primer, 5% v/v dimethylsulfoxide, and an Illustra™ PuReTaq Ready-To-Go™ PCR Bead (GE Healthcare, Pittsburgh, PA, USA) in a final volume of 25 μL. The PCR schedule included an initial denaturation at 94°C for 10 min; 40 cycles of denaturation at 94°C for 30 sec, annealing at 54°C for 30 sec, and extension at 72°C for 1 min; followed by a final extension at 72°C for 10 min. 16S-rDNA amplicons were purified by agarose gel electrophoresis and extracted using a ZymocleanTM Gel DNA Recovery Kit (Zymo Research, Irvine, CA, USA). Sequencing of the amplicons was performed by Eurofins MWG Operon (Louisville, KY). The pA/pH amplicon was sequenced directly using the pA, pH, 16S InL, and 16S InR primers (Table S1). The StrepB/StrepF amplicon was first ligated into a pCR™4-TOPO® vector and cloned into Escherichia coli One ShotTM Top 10 cells using the TOPO® TA Cloning® Kit for Sequencing (Life Technologies, Grand Island, NY, USA). Plasmid DNA from positive transformants was sequenced using T3 and T7 primers (Table S1). Overlapping sequences were assembled into a contig with the CAP3 Sequence Assembly Program [25], available at: http://doua.prabi.fr/software/cap3. Phylogenetic homologies were determined by pairwise nucleotide sequence alignment of the contig using the EZBioCloud tool available at: https://www.ezbiocloud.net/tools/pairAlign. The assembled 16S rDNA sequence was deposited in GenBank, with the accession number MH392705.2.
Studies of PHA depolymerase synthesis
Degradation of PHB or PHBV by Streptomyces sp. SFB5A was assessed by streaking it on SNC agar plates (20 mL) overlaid with 5 mL of SNC agar containing 0.2% w/v of the polymer and observing the formation of clearing zones. To visualize the degradation of PHV during growth, 20 mg of the polymer was dissolved in 1.0 mL of CHCl3 and poured onto the surface of an SNC agar plate. The CHCl3 was allowed to evaporate, forming a thin film of PHV, and 5 mL of molten SNC agar was then carefully poured over the thin film to provide a stable surface for streaking.
For studies of PHA depolymerase synthesis in broth, the organism was grown for 16 h with shaking in NB. Cells were harvested by centrifugation (5,000 × g, 4°C, 10 min) and resuspended in one culture volume of SNC broth containing 20 mM soluble carbon sources (R,S-3HB or R-3HB) or PHA suspensions at 0.2% w/v. Cultures were incubated with shaking. Samples (1.0 to 2.0 mL) were withdrawn at selected times, and clarified by centrifugation. Supernatants were stored at -80°C and analyzed as soon as possible.
Purification of PHA depolymerase
A flask of NB (50 mL) was inoculated with several colonies of Streptomyces sp. SFB5A from a SNC-PHB agar plate culture and incubated with shaking for 16 h. This was used to inoculate two 1 L batches of NB contained in 4 L Erlenmeyer flasks, followed by incubation with shaking for 16 h. Cells were harvested by centrifugation, and the cell pellets were resuspended in two 1 L batches of SNC-PHB broth contained in 4 L Erlenmeyer flasks. The cultures were incubated with shaking for 48 h and then clarified by centrifugation. The supernatant was adjusted to pH 7.4 by adding solid tris(hydroxymethyl)aminomethane (Tris) base, followed by solid ammonium sulfate added to 55% saturation (4°C). Precipitated protein was pelleted by centrifugation (25,000 × g, 20 min, 4°C). The protein pellets were dissolved in 20 mL of Buffer A (25 mM Tris-Cl, 1 mM CaCl2, pH 7.5) and centrifuged to remove particulates (14,000 × g, 10 min, 4°C), followed by addition of 1.17 g of NaCl. This material was loaded onto a 1.5 × 10 cm column of Phenyl Sepharose® CL-4B (GE Healthcare, Wauwatosa, Wisconsin, USA) previously equilibrated with Buffer A containing 1 M NaCl. The column was sequentially washed with 5.6 bed volumes of buffer A containing 1 M NaCl, followed by 7.3 bed volumes of a linear NaCl gradient from 1.0 to 0 M in Buffer A, and 5.6 bed volumes of Buffer A (flow rate 1.2 mL/min, 3.6 mL fractions). Active fractions, eluted during the Buffer A wash, were pooled, concentrated by dialysis against solid sucrose, and applied to a 1.1 × 59 cm column of Sephacryl® S-100-HR (GE Healthcare) previously equilibrated with Buffer A. The column was developed with Buffer A (flow rate 0.1 mL/min, 0.4 mL fractions). Active fractions showing electrophoretic purity were pooled and stored at -80°C. To test for effects of inhibitors and ionic compounds on activity, the enzyme was first dialyzed against 20 volumes of buffer A for 15 h at 4°C with stirring.
PHA depolymerase assays
Routine assays of PHA depolymerase activity were performed turbidometrically at 650 nm [26,27] at 37°C in a 1.0 mL reaction mixture containing 100 mM Tris-HCl, pH 7.9, 1 mM CaCl2, 136 μg of PHB suspension, and enzyme sample (40 to 100 ng purified enzyme or up to 200 μL of culture supernatants). Inhibitors and ionic compounds were added as desired. Activity was calculated using an extinction coefficient at 650 nm of 4.3 μL ng-1cm-1, with a unit defined as 1 μg of PHB degraded per min. Under these conditions, the assay was linear for up to 30 minutes, and the detection limit was about 0.4 units/mL. To test for aPHB degradation, a volume of aPHB suspension was substituted for PHB in the reaction mixture to give the same initial A650. The effect of pH on activity was determined by substituting buffers of overlapping pH for Tris-HCl pH 7.9 in the assay: 100 mM citrate (pH 3-6), piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 6-7.5), Tris-HCl (pH 7-9.5), or glycine-HCl (pH 8.5-10.5).
Qualitative spot tests for PHA depolymerase activity were done as described [27] using 0.1% w/v PHA granule suspensions in 1.0% w/v agarose made up in turbidometric assay buffer. Lipase activity was assessed by the rhodamine B-trioleoylglycerol plate assay [28].
Products resulting from degradation of PHAs were identified by electrospray ionization time-of-flight mass spectrometry (ESI-TOFMS). The reaction mixture contained 180 μg of PHA granules or thin film, 50 mM Tris-HCl (pH 7.9), and 1 mM CaCl2 in a final volume of 1.0 mL. For thin films, before addition of the other ingredients, 180 μL of PHA from a 1% w/v stock in CHCl3 was pipetted into a prewarmed 13 × 100 mm glass screw cap tube, vortexed until the CHCl3 evaporated, and incubated at 50°C for 30 min to drive off residual CHCl3. Purified PHA depolymerase (100 ng) was added to initiate the reactions, followed by incubation for 4 h at 37°C. For time course experiments, the reaction volume was increased from 1 mL to 3 mL, and amounts of all reaction components were tripled accordingly. After incubation, the reaction mixture was acidified to pH 2.5 with HCl. Portions (0.5 mL) were removed to microfuge tubes and centrifuged. The supernatant was extracted three times with 0.5 mL of ethyl acetate, and the organic layer dried under a stream of air at 50°C. The extraction residue was redissolved in 50 μL of 0.1% v/v formic acid and injected at 6 μL/min with a Harvard Laboratories Model 22 syringe infusion pump into an Agilent Technologies 6224 ESI-TOF mass spectrometer operated in negative ionization mode. Nitrogen was used as the desolvation gas at a flow rate of 5 L/h and 325°C, with nebulizer pressure at 30 psig. Fragmentor and skimmer voltages were 175 V and 65 V, respectively. Degradation products were identified from the predicted m/z ratios of the primary ions. A plot of 3HB concentration versus spectral abundance was linear up to at least 48 mM (6,000 ppm) and abundance levels of 2.5 million.
Electrophoretic analyses
Protein purity and subunit Mr were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12% polyacrylamide gels according to Laemmli [29], with silver staining as described [30]; Precision Plus Protein™ Unstained Standards (Bio-Rad Laboratories, Hercules, CA, USA) served as molecular mass markers. Native PAGE was performed using the Laemmli system [29] without SDS or 2-mercaptoethanol. To prepare samples for native PAGE, PHBgrown; cultures were clarified by centrifugation and the supernatants made to 55% saturation (0°C) ammonium sulfate. Precipitated proteins were pelleted by centrifugation (12,000 × g, 10 min, 4°C), and pellets were redissolved in buffer RB containing 1X Laemmli sample buffer [29] without SDS and 2-mercaptoethanol. Dual zymography versus PHB and PHV was done using the same native gel. After electrophoresis of the samples, the gel was overlaid with a slab of 1% agarose (0.75 × 7 × 9 mm) containing 0.1% w/v PHB in turbidometric assay buffer (see above). It was then placed on top of a thin film of PHV previously cast by pipetting 2.5 mL of 1% PHV in CHCl3 evenly onto a (7 × 9 mm) glass plate and allowing the CHCl3 to evaporate. The gel-zymogram assembly was then incubated at 37°C until bands of clearing appeared, indicating PHA depolymerase activity.
Other assays and determinations
Protein concentrations were routinely estimated using the dyebinding assay [31] with bovine serum albumin (BSA) as the standard. For experiments on glucose repression of PHA depolymerase synthesis in broth, glucose concentrations were measured using the dinitrosalicylate assay for reducing sugars [32], with glucose as the standard. Extractable cell protein was determined by pelleting cells from 2.0 ml of culture, washing with deionized water, resuspension in 2.0 mL of 1.0 N NaOH, and heating at 100°C for 10 min. The extracts were clarified by centrifugation, and the protein concentrations in the supernatants were determined using a Bicinchoninic Acid Protein Assay Kit (Sigma-Aldrich) with BSA as the standard. This assay was directly compatible with the NaOH used for extraction.
Cloning and sequencing of the PHA depolymerase gene
Restriction digests, agarose gel electrophoresis, culturing of E. coli , and other routine DNA manipulations were done as described [33]. Cloning of the PHA depolymerase gene (phaZ ) was done using PCRbased techniques; PCR and sequencing primers used are shown in Table S1. Purified PHA depolymerase (90 pmol) was concentrated by trichloroacetic acid precipitation and subjected to SDS-PAGE, followed by blotting onto Mini ProBlott™ polyvinylidene difluoride membranes (Applied Biosystems, Foster City, CA), and staining with Ponceau S according to the manufacturer’s instructions. The PHA depolymerase protein band was excised, and the N-terminal sequence was determined by Edman degradation at the University of Texas Medical Branch Protein Chemistry Laboratory, Galveston, TX, USA. The sequence obtained was: AGLTQVTGFGSNPGNLTMHTYVPDGLAAG. An internal amino acid sequence was similarly obtained by omitting the blotting step after SDS-PAGE and performing in-gel digestion with trypsin before the sequencing procedure, yielding the sequence: SAAPVGTSS.
Degenerate PCR primers PHAD-8 (sense) and PHADIN-3 (antisense) were designed using reverse translations of the amino acid sequences from the N-terminus and internal fragment, respectively. Degeneracy in the sense primer was minimized by using G or C in the Wobble positions of the codons, since the G+C content of Streptomyces species is known to be about 70% [9]. For the antisense primer, a defined base composition of 53.2% G, 38.l% C, 4.6% T, and 4.1% A was incorporated into the wobble positions of the amino acids, based on a codon usage table for Streptomyces coelicolor available at: http://www.kazusa.or.jp/codon/S.html. These primers were used to amplify a 500 base pair (bp) segment (A20TG) of the PHA depolymerase gene. The PCR reaction contained an Illustra PuReTaq™Ready-To-Go™ PCR bead (GE Healthcare, Piscataway, NJ, USA), 118 ng of genomic DNA template, 25 pmol of each primer, and 5% v/v dimethylsulfoxide in a final volume of 25 μL. The PCR schedule included 30 cycles of denaturation (94°C, 1 min), annealing (62.5°C, 30 sec), and extension (72°C, 1 min), followed by an additional extension at 72°C for 10 min.
A combination of PCR-based gene walking techniques was used to obtain additional sequence within and outside of phaZ . These included inverse PCR [34]; single-stranded (ss) DNA ligation PCR [35]; and the method of Siebert et al. [36], using the GenomeWalker™ Universal Kit (Clontech, Mountain View, CA, USA). All procedures were performed essentially as described.
PCR products were purified by agarose gel electrophoresis and extracted from gels using a GenElute™ Gel Extraction Kit (Sigma Aldrich) according to the manufacturer’s instructions. In most cases, the products were first ligated into a pCR®2.1 vector or pCR4®Blunt- TOPO vector (Life Technologies) and introduced into One Shot™ TOP10 E. coli cells. Plasmid DNA from positive E. coli clones was purified using a GenElute™ Plasmid Miniprep Kit (Sigma-Aldrich), and sequenced with M13 Reverse primer (pCR®2.1 clones) or T3 and T7 primers (pCR4®Blunt-TOPO clones) by Eurofins MWG Operon, Louisville, KY, USA. PCR products obtained with the GenomeWalkerTM Universal Kit were sequenced directly with primers GSP2 and AP2.
As a final check, the entire phaZ coding sequence was amplified from genomic DNA by PCR with primers PHAZF9 and PHAZR9, which contained engineered BamHI and HindIII restriction sites, respectively. The PCR product was digested with BamHI and HindIII and ligated with similarly digested plasmid pUC18 [31]. The resulting construct was cloned into E. coli DH5α, and plasmid DNA extracted from positive transformants was sequenced with M13 forward and reverse primers plus internal, phaZ -specific primers (PHAZL3, PHAZInF, PHAZInR, and PHAZR4). The nucleotide sequence of phaZ was deposited in GenBank, with the accession number MH746934.
Sequence analysis
Sequences from phaZ were assembled with the CAP3 Sequence Assembly Program [25], and open reading frames identified with the ExPASy Translate utility (https:// web.expasy.org/translate/). Initial homology searches of the predicted amino acid sequence were done with the Protein BLAST tool (https:// blast.ncbi. nlm.nih.gov/Blast.cgi) against the RefSeq protein database. To create a multiple sequence alignment, the predicted amino acid sequence was first searched with Protein BLAST against the PHA Depolymerase Engineering Database http:// www. ded.uni-stuttgart.de/ [4]. Eight sequences with the best homology scores were aligned with the Streptomyces sp. SFB5A deduced amino acid sequence using the T-Coffee program (http://tcoffee.vital-it.ch/apps/tcoffee/do:regular) [37]. A potential signal peptide was located using Predisi (http: //www.predisi.de/) [38]. A model of the three-dimensional structure of the PHA depolymerase was constructed from its deduced amino acid sequence using PHYRE2 [39], available at: http:// www. sbg.bio.ic.ac.uk/phyre2/html/page.cgi? id=index.
Results
Isolation and characterization of Streptomyces sp. SFB5A
Isolate SFB5A formed filamentous, dry, flat, and opaque colonies after 5 d of incubation on SNC-agar plates containing PHB, glucose, or other carbon sources, and emitted the odor of geosmins characteristic of actinomycetes. A substrate mycelium was formed during the first 24 to 36 h of incubation, followed by a powdery white aerial mycelium. After 3 to 5 d, colonies became grey-brown, indicating sporulation, with color more pronounced at the periphery than in the center. Gram stains showed a mixture of Gram positive branched filaments, rounded rods approximately 0.5 × 1.0 μm, and spores in older cultures. A soluble, brown pigment was formed after 3 to 5 d of growth on TSA, MSF, or SNC plus glucose agar, but not on SNC-PHB agar. Colonies on complex agar media such as TSA and NA were moister and less filamentous than on SNC-PHB agar, wrinkled, off-white, with undulate margins, and failed to exhibit sporulation even after incubation for over 3 weeks. Growth in SNC broth media was very flocculent, and cells adhered to the sides and bottom of shake flasks. In contrast, growth in complex media such as NB or TSB was much faster and more dispersed than in SNC broth. The growth rate and doubling time in NB at 30°C, estimated by turbidometry, were 0.75 h-1 and 56 min, respectively. The temperature range for growth on TSA plates was 15-39°C, with optimal growth between 30 and 35°C.
BLAST searches of a 1324 bp portion of the 16S rRNA gene amplified from isolate SFB5A showed the highest identity to 16S rRNA from Streptomyces cinereoruber subsp. cinereoruber NBRC 12756 [Accession #AB184121; 99.62% similarity (1319 bp), 99.7% completeness]. However, preliminary results of physiological tests and morphological comparisons between the two strains suggested that they may represent separate species, and further taxonomic identification is in progress. Therefore, the isolate was designated as Streptomyces sp. SFB5A.
Degradation of PHA in cultures
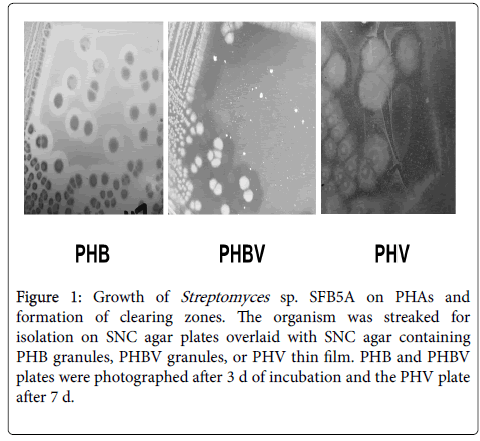
Streptomyces sp. SFB5A grew on SNC agar plate overlays containing PHB or PHBV. Clearing zones appeared after about 30 to 48 h, during the transition from vegetative to aerial mycelium, with full clearing after 3 to 5 d (Figure 1). It also grew slowly on SNC agar plates overlaid with PHV thin films, and full clearing of the PHV required up to 7 d (Figure 1).
Figure 1: Growth of Streptomyces sp. SFB5A on PHAs and formation of clearing zones. The organism was streaked for isolation on SNC agar plates overlaid with SNC agar containing PHB granules, PHBV granules, or PHV thin film. PHB and PHBV plates were photographed after 3 d of incubation and the PHV plate after 7 d.
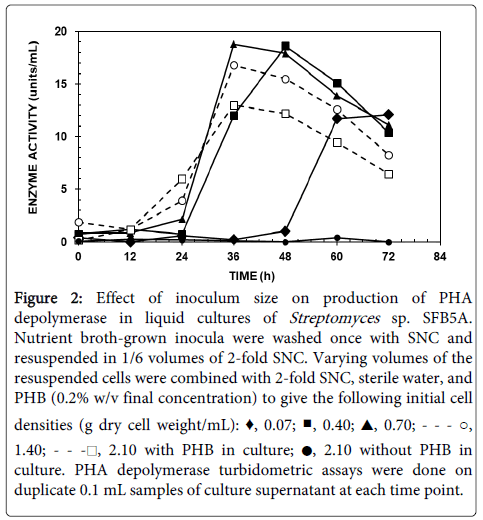
When NB-grown cells were resuspended in SNC-PHB broth and incubated in shake culture, PHA depolymerase activity was detected after a lag of 12 to 24 h, peaking at 36 to 48 h, and declining rapidly thereafter, possibly because of proteolytic degradation (Figure 2). Maximal activity levels were obtained with inocula containing 0.4 to 0.7 mg of dry cell weight/mL.
Figure 2:Effect of inoculum size on production of PHA depolymerase in liquid cultures of Streptomyces sp. SFB5A. Nutrient broth-grown inocula were washed once with SNC and resuspended in 1/6 volumes of 2-fold SNC. Varying volumes of the resuspended cells were combined with 2-fold SNC, sterile water, and PHB (0.2% w/v final concentration) to give the following initial cell densities (g dry cell weight/mL): ♦, 0.07; ■, 0.40; ▲, 0.70; - - - ○, 1.40; - - -□, 2.10 with PHB in culture; ●, 2.10 without PHB in culture. PHA depolymerase turbidometric assays were done on duplicate 0.1 mL samples of culture supernatant at each time point.
The latter corresponded to resuspension of pelleted NB-grown cells in a volume of SNC-PHB broth equal to that of the original culture, so this inoculum level was used for subsequent experiments on PHA depolymerase production in broth. The lag time with inocula that were washed was about 12 h longer than that for unwashed inocula, although the maximal PHA depolymerase activities observed were similar (not shown). Use of log phase cells versus stationary phase cells as inocula decreased the maximal activity observed by about 3-fold but did not affect the lag time (not shown).
Activity was also obtained in SNC broth cultures containing R,S-3HB, R-3HB, or PHBV but less than with PHB (26.8%, 14.7%, and 4.9% respectively of activity with PHB). Activity from broth containing PHV or lacking any carbon source was at or below the detection limit for the assay. PHA depolymerase synthesis in broth cultures was repressed by glucose and resumed after exhaustion of glucose from the medium (Figure S1). A similar repression phenomenon was observed in agar cultures containing both PHB and 0.5% glucose, clearing zones requiring 4 to 5 d to appear instead of 2 d with PHB alone (not shown). Use of PHB plus 1% glucose increased this time to at least 7 d.
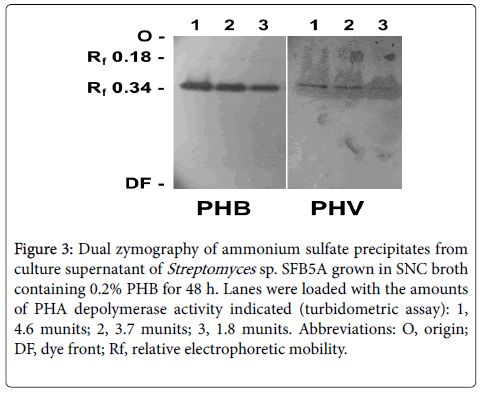
Dual zymography revealed that a PHA depolymerase active against both PHB and PHV was produced during growth in SNC-PHB broth (Figure 3). Identical activity bands of Rf 0.34 appeared on both the PHB and PHV zymograms. A faint band at Rf 0.18 also appeared on the PHB zymogram but not on the PHV zymogram.
Figure 3: Dual zymography of ammonium sulfate precipitates from culture supernatant of Streptomyces sp. SFB5A grown in SNC broth containing 0.2% PHB for 48 h. Lanes were loaded with the amounts of PHA depolymerase activity indicated (turbidometric assay): 1, 4.6 munits; 2, 3.7 munits; 3, 1.8 munits. Abbreviations: O, origin; DF, dye front; Rf, relative electrophoretic mobility.
Purification and characterization of PHA depolymerase
PHA depolymerase was purified 186-fold to approximately 97% electrophoretic homogeneity from PHB-grown culture supernatants of Streptomyces sp. 5A, with a final yield of 11 μg protein and 16% recovery of activity. Its subunit Mr was approximately 47,000 (Figure S2). It was stable during storage for 4 d at 25°C, 7 d at 4°C, at least 14 d at -20°C, and indefinitely at -80°C. It lost only 10% of its original activity after 5 cycles of freezing at -80°C and thawing at 20°C. Optimal activity was obtained from pH 7.0 to 8.5, and at 45°C.
The effects of ionic compounds were tested (Table 1). CaCl2, MgCl2, and KCl stimulated activity, probably because of the cationic moieties since NaCl had no effect on activity. NaNO3, ZnCl2, Na2SO4, and NH4Cl strongly inhibited activity, while Na2CO3 and Na2HPO4 were slightly inhibitory. The effects of common enzyme inhibitors on PHA depolymerase activity were also tested (Table 1). The enzyme was highly inhibited by the sulfhydryl reducing agent, dithiothreitol (DTT), but only weakly by the sulfhydryl alkylation reagent, iodoacetate (IAA). It was not inhibited by KCN. The non-ionic detergents Triton X-100 and Tween-20 completely inhibited activity. Activity was strongly inhibited by the divalent ion chelator, ethylenediaminetetraacetic acid (EDTA), but was restored by addition of 5 mM CaCl2 to the assay mixture, confirming the stimulatory effect of CaCl2. Superficially, the serine hydrolase inhibitor, phenylmethylsulfonyl fluoride (PMSF) inhibited activity at 1 mM, but ethanol used to prepare the PMSF stock solution also inhibited activity so that the net inhibition was only 2.5%. The enzyme lacked lipase activity (not shown), as determined by a specific plate assay [28].
| Ionic Compounds | ||
|---|---|---|
| Compounda | Final concentration (mM) | % of Controle |
| Deionized water (no CaCl2) | --- | 100 |
| CaCl2 | 2 | 245 |
| MgCl2 | 2 | 188 |
| ZnCl2 | 2 | 33.3 |
| NaCl | 4 | 101 |
| Na2CO3 | 2 | 82.3 |
| Na2HPO4 | 2 | 83.3 |
| Na2SO4 | 2 | 45.8 |
| KCl | 4 | 117 |
| NH4Cl | 4 | 54.2 |
| NaNO3 | 4 | <0 |
| Enzyme Inhibitors | ||
| Deionized water (plus CaCl2) | --- | 100 |
| DTT | 1 | 30.4 |
| EDTA | 1 | 6.1 |
| EDTA plus 5 mM CaCl2b | 1 | 136 |
| KCN | 1 | 96.6 |
| PMSFc | 1 | 66.4 |
| Ethanolc | 856 | 68.9 |
| IAA | 1 | 82.6 |
| Triton X-100 | ---d | <0 |
| Tween 20 | ---d | <0 |
Table 1: Effects of selected ionic compounds and common enzyme inhibitors on activity of PhaZSsp5A. aThe standard turbidometric assay reaction mixture was supplemented with the test compounds at the concentrations indicated. For testing of ionic compounds, CaCl2 was omitted from the standard assay mixture. For testing of common enzyme inhibitors, the standard assay mixture (i.e., containing 1.0 mM CaCl2) was used. bCaCl2 was added to a final concentration of 5.0 mM in this assay mixture. c Since the PMSF stock was made up in ethanol, a control containing an equivalent volume of ethanol (50 ml per mL of assay mixture) was done. The final concentration of ethanol was thus ~856 mM final concentration. dThe concentrations of Triton X-100 and Tween 20 were 0.1% v/v. ePercentage of activity versus deionized water control without CaCl2 (for ionic compounds) or with CaCl2 (for enzyme inhibitors). Values are averages from triplicate assays. Activities were corrected for absorbance changes obtained with minus enzyme controls. Values below zero indicate that final absorbance readings were slightly above that of the minus enzyme control.
Qualitative spot tests indicated that the purified PHA depolymerase degraded suspensions of PHB, PHBV, and PHV (not shown), although formation of clearing zones on PHV was much slower and less complete than with the other two PHAs. The enzyme did not degrade aPHB suspensions as assayed turbidometrically. The products from PHA degradation were identified by ESI-TOF-MS (Table 2; see Figure S3 for representative mass spectra). The enzyme degraded PHB granules and thin films to mostly 3HB monomer, with small and variable amounts of 3HB dimer and trimer; higher order oligomers were not detected. Controls without enzyme showed no significant product formation after 4 h (not shown). The enzyme also degraded granules and thin films of PHBV to primarily the monomers of 3HB and 3HV, with smaller amounts of the dimer and trimer forms. Furthermore, the enzyme degraded PHV granules and thin films, with a similar product pattern of 3HV units. The enzyme did not significantly degrade PHO thin films; total spectral abundance of 3HO obtained was only about 0.9% of total products obtained with PHV thin films.
| PHA Substratea | Percent of totalb | Total Abundancec | ||||||
|---|---|---|---|---|---|---|---|---|
| 3HB Forms | 3HV Forms | 3HO | ||||||
| 1-mer | 2-mer | 3-mer | 1-mer | 2-mer | 3-mer | 1-mer | ||
| PHB-G | 80.8 | 11.3 | 7.9 | - | - | - | - | 9,78,272 |
| PHB-TF | 82.1 | 10.3 | 7.6 | - | - | - | - | 10,60,839 |
| PHV-G | - | - | - | 72.8 | 13.8 | 13.4 | - | 9,64,429 |
| PHV-TF | - | - | - | 77.6 | 12.9 | 9.5 | - | 8,37,957 |
| PHBV-G | 49.8 | 5.7 | 3.8 | 37.3 | 2.9 | 0.6 | - | 22,65,534 |
| PHBV-TF | 50.5 | 4.9 | 2.1 | 39.1 | 3.1 | 0.4 | - | 22,23,732 |
| PHO-TF | - | - | - | - | - | - | 100 | 7588 |
Table 2: Pattern of products from degradation of various PHAs by PhaZSsp5A. aPHAs (180 µg) in granular (G) or thin film (TF) form were incubated with 100 ng purified PHA depolymerase for 4 h and processed for ESI-TOF-MS as described in Materials and Methods. bData indicate the percentage of total spectral abundance for the forms indicated and are averages from at least two replicates for each PHA tested. Values were corrected for abundances of each form obtained with controls lacking enzyme, which were at or near the background level for the instrument (abundance level 10,000). -, no product detected. cObtained by totaling the spectral abundances of all the products indicated for each PHA.
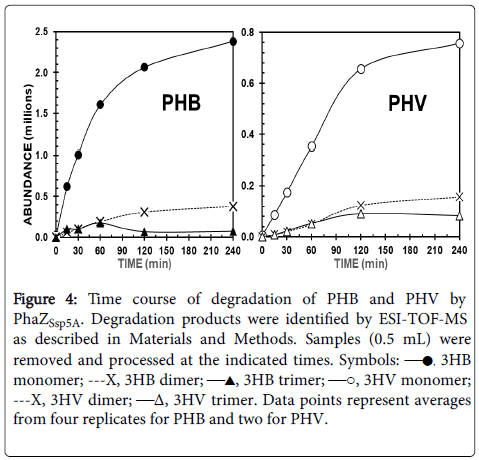
Time courses of PHB and PHV degradation to monomers and dimers were roughly hyperbolic (Figure 4). The enzyme degraded PHB 9.7 times faster than PHV, based on rates calculated from the formation of total products in the first 15 min, and generated 2.1 times more products from PHB versus PHV after 4 h. With PHB as substrate, production of 3HB trimers peaked at 60 min and then decreased, whereas production of 3HB dimers increased steadily and then stabilized. A similar pattern was obtained with PHV as substrate, although production of 3HV trimers peaked only after 120 min, and not as sharply as with PHB.
Figure 4: Time course of degradation of PHB and PHV by PhaZSsp5A. Degradation products were identified by ESI-TOF-MS as described in Materials and Methods. Samples (0.5 mL) were removed and processed at the indicated times. Symbols: —●, 3HB monomer; ---X, 3HB dimer; —▲, 3HB trimer; —○, 3HV monomer; ---X, 3HV dimer; —Δ, 3HV trimer. Data points represent averages from four replicates for PHB and two for PHV.
Cloning of the PHA depolymerase gene
Attempts to clone DNA directly from Streptomyces sp. SFB5A into E. coli , including preparation of partial and total gene libraries, were unsuccessful. Therefore, phaZ was cloned by using PCR-based methods, as illustrated in Figure S4. A 500 bp segment of phaZ was obtained by using PCR with degenerate, gene-specific primers designed using amino acid sequence information from purified PHA depolymerase protein. Cloning of the rest of the gene was accomplished with three different PCR-based gene walking techniques.
An open reading frame (ORF) of 1505 bp was ultimately obtained after sequence assembly, encoding a protein containing 501 amino acids and molecular weight of 51,377 Daltons. The ORF contained 71.8% G+C residues, consistent with the high G+C content of streptomycetes [9]. Analysis of the open reading frame with Predisi [38] revealed a putative signal peptide containing 48 amino acids. This was followed by sequence encoding a peptide of 453 amino acids, with a calculated isoelectric point of 4.83 and molecular weight of 46,400 Daltons, which corresponded well with the Mr of the purified PHA depolymerase (47,000) determined by SDS-PAGE (Figure S2). In addition, the first 29 amino acids of this predicted peptide agreed exactly with those obtained from N-terminal amino acid sequencing of purified PHA depolymerase (see Materials and Methods). Initial Protein BLAST searches of the predicted amino acid sequence revealed significant homology to esterases and PHA depolymerases from Streptomyces and other actinomycetes.
Discussion
We describe here the isolation of Streptomyces sp. SFB5A, which degraded PHB, PHV, and PHBV in agar overlays and broth culture. PHA depolymerase activity, assayed turbidometrically with PHB as substrate, was detected in culture supernatants from growth of the organism in liquid medium containing PHB, PHBV, or 3HB, but not PHV. Interestingly, PHV was degraded slowly in agar overlay cultures and by purified enzyme (see below), but PHA depolymerase activity was not detected in broth cultures containing PHV, even after extended incubation. Perhaps the enzyme adhered tightly to PHV in broth culture, so that it would not be detectable in culture supernatants. As observed with many other extracellular PHA depolymerases [2,5-7], synthesis of PHA depolymerase was repressed in liquid and agar media containing both glucose and PHB until glucose was exhausted. Zymography suggested that only one PHA depolymerase was produced by this isolate during growth on PHB and that the enzyme could degrade both PHB and PHV. However, we did not entirely rule out the possibility of multiple PHA depolymerases with the same or different electrophoretic mobility on native gels. The faint Rf 0.18 band on the PHB zymogram (Figure 3) could thus represent a different isoform of PHA depolymerase, but more likely self-aggregates or complexes with other proteins.
A PHA depolymerase (heretofore abbreviated PhaZSsp5A) was purified from liquid cultures of Streptomyces sp. SFB5A containing PHB. Combined results of qualitative spot tests, turbidometric assays, and mass spectrometry showed that the purified enzyme could degrade PHB, PHV, and PHBV, but not PHO or aPHB, placing it in the family of extracellular denatured PHASCL depolymerases [2]. It shared several characteristics with extracellular dPHASCL depolymerases from other species of Streptomyces (Table 3), and PHA depolymerases in general [2]. These included an alkaline pH optimum (with the exception of S. ascomycinicus ), temperature optimum of 40-60°C, high stability, monomer structure of approximately 40-60 kDa, affinity for hydrophobic interaction chromatography resins, and stimulation by divalent cations. Notably, activity was strongly inhibited by NH4+ and SO4 -2, which necessitated careful removal of excess (NH4)2SO4 from pellets after the ammonium sulfate precipitation step of purification prior to assay. The strong inhibition by the sulfhydryl reducing agent, DTT, but weak inhibition by the sulfhydryl alkylation reagent, IAA, suggested that disulfide linkages but not free sulfhydryl groups were crucial for activity. Lack of inhibition with KCN indicated that iron or other susceptible metals were not involved in catalysis. Non-ionic detergents strongly inhibited its activity, possibly by interfering with binding of the relatively hydrophobic PHB to its binding site within the enzyme. The enzyme was not significantly inhibited by the serine hydrolase inhibitor, PMSF, known to inhibit some PHA depolymerases [2].
Mass spectral analysis showed that the purified enzyme degraded PHB, PHV, and PHBV in either granule or thin film form to a mixture of the corresponding monomers, dimers, and trimers, with monomers as the predominant product (Table 2). Since higher order oligomers were not detected, this pattern suggested an exolytic rather than an endolytic cleavage mechanism. The pattern of products arising from degradation of PHB by extracellular dPHASCL depolymerases is variable. Some produce only 3HB monomer [5,40], others produce various proportions of monomer and dimer [6,14,41,42], while still others produce mostly dimer with smaller amounts of monomer and/or trimer [2,43,44]. The enzyme from Comomonas acidovorans forms different proportions of degradation products from PHB depending on enzyme concentration, with mostly dimer and smaller amounts of monomer formed at low enzyme concentrations (0.2 μg/mL) and the opposite proportions at high enzyme concentrations (20 μg/mL) [6].
However, most reports of PHA degradation by PHA depolymerases only state one-point composition of the products after several hours of incubation. In contrast, we used ESI-TOF-MS to monitor the change in product composition over time, which might better reveal information about the catalytic mechanism. Time courses of PHB and PHV degradation by PhaZSsp5A (Figure 4) showed that formation of the corresponding monomers and dimers was essentially hyperbolic, possibly due to substrate depletion or product inhibition. In thin film reaction mixtures, remnants of the films were still visible after 4 h, suggesting the latter as a more likely possibility. PHB, PHBV, and PHV yielded comparable patterns of degradation products (Table 2) suggesting that the catalytic mechanisms for degradation of these PHAs were similar. However, qualitative spot tests showed that PHB was degraded faster than PHV, indicating that PHB was a preferred substrate for the enzyme. Indeed, mass spectral assays showed that the initial rate of monomer formation from PHB was considerably greater than that from PHV, and the product yields from PHV at 4 h were about 2-fold lower. The slower degradation of PHV versus PHB could be because the 3HV monomer has one more carbon in its side chain than 3HB, resulting in greater steric hindrance or hydrophobic effects at the catalytic site of the enzym
The deduced amino acid sequence of PhaZSsp5A included a putative 48 amino acid signal peptide of 48 amino acids containing the sequence: RRWLT. This is similar to the twin-arginine translocation (Tat) signature found in Streptomyces coelicolor : RRXϕϕ (where ϕ is a hydrophobic amino acid); the Tat pathway has been identified as a major translocation system for secreted proteins in Streptomyces coelicolor [45].
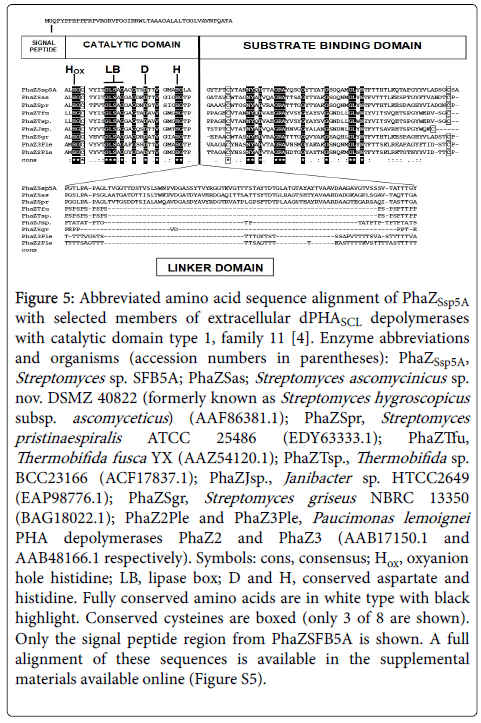
A BLAST search of the PhaZSsp5A deduced amino acid sequence versus the PHA Depolymerase Engineering Database [4] revealed the highest homology to extracellular dPHASCL depolymerases with catalytic domain type 1, family 11. The best homology was with the PHA depolymerase of Streptomyces ascomycinicus (51% identity with 98% coverage). An abbreviated alignment with eight selected matches from this group is shown (Figure 5; Figure S5 shows the complete alignment). The putative signal peptide of PhaZSsp5A was followed by a type 1 catalytic domain [2]. This contained a triad of strictly conserved amino acids (numbers indicate the reside number in the secreted PhaZSsp5A, i.e., excluding the signal peptide): an oxyanion hole histidine (H85), a serine (S168) contained within a lipase box (G-L-SA- G), as well as an aspartate (D181). Site-directed mutagenesis reveals that these three amino acids are necessary for catalysis (reviewed in reference 2). PhaZSsp5A also had a second conserved histidine (H305) found in the catalytic domain of PHB depolymerases [2]. A putative type 1 substrate binding domain [2] occurred at the C-terminal end of the predicted amino acid sequence of PhaZSsp5A. This contained several conserved hydrophobic and other amino acids, notably several tyrosine residues. In addition, PhaZSsp5A also featured eight cysteine residues, six of which were located in the catalytic domain (C87, C125, C198, C208, C318, and C330) and the other two (C443 and C499) in the substrate binding domain (Figure S5). The positions of these cysteines were highly conserved among the nine enzymes aligned and are likely involved in disulfide bond formation, which would explain the strong inhibition of PhaZSsp5A activity by DTT. A putative linker domain with a fibronectin type III (Fn3) signature [2] was located between the catalytic domain and substrate binding domain of PhaZSsp5A. However, only PhaZSsp5A and the enzymes from Streptomyces ascomycinicus and Streptomyces pristinaespiralis contained the Fn3 signature among those compared; the six others contained threonine-rich regions [2] instead. Further research might reveal whether the type of linker domain in these enzymes influences their reaction mechanisms and composition of their PHA degradation products.
Figure 5: Abbreviated amino acid sequence alignment of PhaZSsp5A with selected members of extracellular dPHASCL depolymerases with catalytic domain type 1, family 11 [4]. Enzyme abbreviations and organisms (accession numbers in parentheses): PhaZSsp5A, Streptomyces sp. SFB5A; PhaZSas; Streptomyces ascomycinicus sp. nov. DSMZ 40822 (formerly known as Streptomyces hygroscopicus subsp. ascomyceticus ) (AAF86381.1); PhaZSpr, Streptomyces pristinaespiralis ATCC 25486 (EDY63333.1); PhaZTfu, Thermobifida fusca YX (AAZ54120.1); PhaZTsp., Thermobifida sp. BCC23166 (ACF17837.1); PhaZJsp., Janibacter sp. HTCC2649 (EAP98776.1); PhaZSgr, Streptomyces griseus NBRC 13350 (BAG18022.1); PhaZ2Ple and PhaZ3Ple, Paucimonas lemoignei PHA depolymerases PhaZ2 and PhaZ3 (AAB17150.1 and AAB48166.1 respectively). Symbols: cons, consensus; Hox, oxyanion hole histidine; LB, lipase box; D and H, conserved aspartate and histidine. Fully conserved amino acids are in white type with black highlight. Conserved cysteines are boxed (only 3 of 8 are shown). Only the signal peptide region from PhaZSFB5A is shown. A full alignment of these sequences is available in the supplemental materials available online (Figure S5).
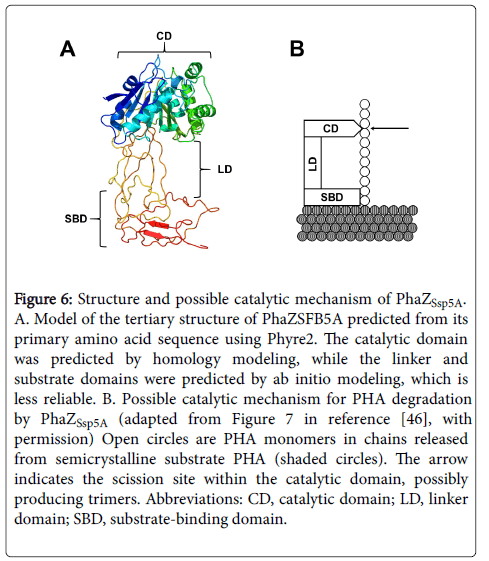
In silico modeling of the PhaZSsp5A tertiary structure revealed a large cleft between the catalytic and substrate-binding domains, bridged by the Fn3-like linker domain (Figure 6). The linker domain is proposed to properly juxtapose the catalytic and substrate binding domains [2]. Site-directed mutagenesis studies of the extracellular dPHB depolymerase of Ralstonia pickettii T1 suggest that its substratebinding domain adsorbs to and disrupts the semi-crystalline PHB surface, solubilizing PHB chains and making them available to the catalytic domain for hydrolysis [46]. We propose that the length of the linker domain may constrain the length of the solubilized chain available to the scission site and thus influence the length of the initial degradation product. The peaking of trimer levels during PHA degradation by PhaZSsp5A (Figure 4) suggests that they were an initial product of PHA degradation, perhaps due to this constraint, as modeled in Figure 6. Once trimers were formed, they could have reentered the catalytic site by diffusion and been cleaved randomly to monomers or dimers, both of which reached a steady state (Figure 4). Alternatively, the length of the initial product could have been determined by the monomer-binding capacity of the catalytic site. For example, Ohura et al. demonstrate that the PHB depolymerase of Pseudomonas stutzeri preferentially binds three 3HB units, although it recognizes at least two 3HB units for hydrolysis [47].
Figure 6: Structure and possible catalytic mechanism of PhaZSsp5A. A. Model of the tertiary structure of PhaZSFB5A predicted from its primary amino acid sequence using Phyre2. The catalytic domain was predicted by homology modeling, while the linker and substrate domains were predicted by ab initio modeling, which is less reliable. B. Possible catalytic mechanism for PHA degradation by PhaZSsp5A (adapted from Figure 7 in reference [44], with permission) Open circles are PHA monomers in chains released from semicrystalline substrate PHA (shaded circles). The arrow indicates the scission site within the catalytic domain, possibly producing trimers. Abbreviations: CD, catalytic domain; LD, linker domain; SBD, substrate-binding domain.
Crystallization of wild type and mutated versions of enzymes is useful for elucidating their substrate-binding and catalytic mechanisms. Only two extracellular PHA depolymerases have been crystallized to date, both of which are considerably different from PhaZSsp5A: the dPHB depolymerase from Penicillium (Talaromyces) funiculosum [48,49], and the PhaZ7 enzyme from Paucimonas lemoignei [50,51]. The Penicillium enzyme has a type 2 rather than a type 1 catalytic domain, is considerably smaller than PhaZSsp5A (33,000 Daltons versus 46,400), and lacks the typical linker and substratebinding domains described above [49]. Its PHA binding site is identified as a crevice on its surface, with several hydrophobic residues and three or possibly four monomer binding subsites. The catalytic triad residues (S39, D121, and H155) are clustered at the mouth of the crevice. Crystallized muteins of PhaZ7 reveal five amino acids (Y105, Y176, Y189, Y190, and W207) located in a cleft shown to be necessary for binding of 3HB tetramer [51]. Unlike PhaZSsp5A, PhaZ7 is specific for nPHB and aPHB, rather than dPHB. However, nine tyrosine residues occurred in the substrate-binding domain of PhaZSsp5A, and were well conserved among the PHA depolymerases compared in Figure 5. By analogy, these could be involved in PHA substrate binding.
We are currently attempting to overexpress phaZ in a heterologous host and thereby obtain sufficient quantities of PhaZSsp5A for crystallization. This will allow us to better study the catalytic, linker, and substrate domains of the enzyme and to clarify the reaction mechanism of the enzyme.
Acknowledgments
We thank Dr. Douglas Dennis for piquing our interest in PHAs. We are very grateful to Dr. Christine Hughey of the James Madison University Chemistry Department for assistance with ESI-TOF-MS. Funding was provided by grant #J-713 from the Thomas F. and Kate Miller Jeffress Memorial Trust; the National Science Foundation (NSF) Research Experience for Undergraduates program (awards DBI-0649045, CHE-0097448, DMR-0097449, and CHE-1461175); an NSF Research Opportunity Award (supplement to award MCB-9514100 to Dr. Douglas Dennis); and the American Chemical Society Petroleum Research Fund Type B #33989-B5-SF01. We also acknowledge funding from Bridgewater College via the Research Experience at Bridgewater, Faculty Research Grants, the Harry G.M. Jopson Endowment, and the Frances E. Silliman Research Endowment. We thank Dr. Bradley Stiles for critical reading of the manuscript.
References
- Anderson AJ, Dawes EA (1990) Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiological Reviews 54: 450-472.
- Jendrossek D, Handrick R (2002) Microbial degradation of polyhydroxyalkanoates. Annu Rev Microbiol 56: 403-432.
- Horowitz DM, Sanders KM (1994) Amorphous, biomimetic granules of polyhydroxybuyrate: preparation, characterization, and biological implications. J Am Chem Soc 116: 2695-2702.
- Knoll M, Hamm TM, Wagner F, Martinez V, Pleiss J (2009) The PHA Depolymerase Engineering Database: A systematic analysis tool for the diverse family of polyhydroxyalkanoate (PHA) depolymerases. BMC Bioinformatics 10: 89.
- Jendrossek D, Knoke I, Habibian RB, Steinbuchel A, Schlegel HG (1993) Degradation of poly(3-hydroxybutyrate), PHB, by bacteria and purification of a novel PHB depolymerase from Comamonas sp. J Environ Polymer Degradation 1: 53-63.
- Kasuya K, Inoue Y, Tanaka T, Akeheta T, Iwata T, et al. (1997) Biochemical and molecular characterization of the polyhydroxybutyrate depolymerase of Comamonas acidovorans YM1609, isolated from freshwater. Appl Environ Microbiol 63: 4844-4852.
- Klingbeil B, Kroppenstedt RM, Jendrossek D (1996) Taxonomic identification of Streptomyces exfoliatus K10 and characterization of its poly(3-hydroxybutyrate) depolymerase gene. FEMS Microbiol Lett 142: 215-221.
- Schirmer A, Jendrossek D, Schlegel HG (1993) Degradation of poly(3-hydroxyoctanoic acid (P(3HO)) by bacteria. Purification and properties of a P(3HO) depolymerase from Pseudomonas fluorescens GK13. Appl Environ Microbiol 59: 1220-1227.
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces Genetics. John Innes Foundation, Norwich, UK.
- Peczynska-Czoch W, Mordarski M (1988) Actinomycete enzymes. Actinomycetes in Biotechnology. Academic Press, London, UK.
- Mergaert JA, Webb A, Anderson C, Wouters A, Swings J (1993) Microbial degradation of poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in soils. Appl Environ Microbiol 59: 3233-3238.
- Aly MM, Tork S, Qari HA, Al-Seeni MN (2015) Poly-ß-hydroxybutyrate depolymerase from Streptomyces lydicus MM10, isolated from wastewater sample. Int J Agricul Biol 17: 891-900
- Calabia BP, Tokiwa Y (2006) A novel PHB depolymerase from a thermophilic Streptomyces sp. Biotechnol Lett 28: 383-388.
- Garcia-Hidalgo J, Hormigo D, Arroyo M, de la Mata I (2013) Novel extracellular PHB depolymerase from Streptomyces ascomycinicus: PHB copolymers degradation in acidic conditions. PLos ONE 8: e71699.
- Hsu KJ, Tseng M, Don TM, Yang MK (2012) Biodegradation of poly(ß-hydroxybutyrate) by a novel isolate of Streptomyces bangladeshensis 77T-4. Botanical Studies 53: 307-313.
- Gangoiti J, Santos M, Prieto MA, de la Mata I, Serra JL, et al. (2012) Characterization of a novel subgroup of extracellular medium-chain-length polyhydroxyalkanoate depolymerases from actinobacteria. Appl Environ Microbiol 78: 7229-7237.
- Kim HJ, Kim DY, Nam JS, Bae KS, Rhee YH (2003) Characterization of an extracellular medium-chain-length poly(3-hydroxyalkanoate) depolymerase from Streptomyces sp. KJ-72. Antonie van Leeuwenhoek 83: 183-189.
- Santos M, Gangoiti J, Keul H, Moller M, Serra JL, et al. (2013) Polyester hydrolytic and synthetic activity catalyzed by the medium-chain-length poly(3-hydroxyalkanoate) depolymerase from Streptomyces venezuelae SO1. Appl Microbiol Biotechnol 97: 211-222.
- Steinbuchel A, Schmack G (1995) Large-scale production of poly(3-hydroxyvaleric acid) by fermentation of Chromobacterium violaceum, processing, and characterization of the homopolyester. J Environ Polymer Degradation 3: 243-258.
- Timm A, Steinbuchel A (1990) Formation of polyesters of medium-chain-length 3-hydroxyalkanoic acids from gluconate by Pseudomonas aeruginosa and other fluorescent pseudomonads. Appl Environ Microbiol 56: 3360-3367.
- Schlegel HG, Kaltwasser H, Gottschalk G (1961) A submersion method for culture of hydrogen-oxidizing bacteria: growth physiological studies. Arch Mikrobiol 38: 209-222.
- Pospiech A, Neumann B (1995) A versatile quick-prep of genomic DNA from gram positive bacteria. Trends Genet 11: 217-218.
-  Rintala H, Nevalainen A, Ronka E, Suutari M (2001) PCR primers targeting the 16S rRNA gene for the specific detection of      streptomycetes.Mol Cell Probes 15: 337-347
- Edwards U, Rogall T, Blocker H, Emde M, Bottger E (1989) Isolation and direct complete nucleotide determination of entire genes.Characterization of a gene coding for 16S ribosomal RNA. Nucl Acids Res 17: 7843-7853.
- Huang X, Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9: 868-877.
- Jendrossek D, Frisse A, Behrends A, Andermann M, Kratzin HD, et al. (1995) Biochemical and Molecular Characterization of the Pseudomonas lemoignei Polyhydroxyalkanoate Depolymerase System. J Bacteriol 177: 596-607.
- Jendrossek D, Muller B, Schlegel HG (1993) Cloning and characterization of the poly(hydroxyalkanoic acid)-depolymerase gene locus, phaZ1, of Pseudomonas lemoignei and its gene product. Eur J Biochem 218: 701-710.
- Kouker G, Jaeger KE (1987) Specific and sensitive plate assay for bacterial lipases. Appl Environ Microbiol 53: 211-213.
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680-685.
- Bassam BJ, Caetano-Anolles G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196: 80-83.
- Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31: 426-428.
- Bradford MM (1976) Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254.
- Ausubel F, Brent R, Kingston R, Moore D, Seidmann J, et al. (1990) Short Protocols in Molecular Biology. Wiley-Interscience, New York, USA.
- Ochman H, Gerber A, Hartl D (1988) Genetic applications of an inverse polymerase chain reaction. Genetics 120: 621-623.
- Zhang XH, Chiang V (1996) Single-stranded DNA ligation by T4 RNA ligase for PCR cloning of 5’-noncoding fragments and coding sequence of a specific gene. Nucl Acids Res 24: 990-991.
- Siebert P, Chenchik A, Kellogg D, Lukyanov K, Lukyanov S (1995) An improved PCR method for walking in uncloned genomic DNA. Nucl Acids Res 23: 1087-1088.
- Notredame C, Higgins DG, Heringa J (2000) T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205-217.
- Hiller K, Grote A, Scheer M, Munch R, Jahn D (2004) PrediSi: prediction of signal peptides and their cleavage positions. Nucl Acids Res 32: W375-W379.
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 10: 845-858.
- Yamada K, Mukai K, Doi Y (1993) Enzymatic degradation of poly(hydroxyalkanoates) by Pseudomonas pickettii. Int J Biol Macromol 15: 215-220.
- Garcia-Hidalgo J, Hormigo D, Prieto MA, Arroyo M, de la Mata I (2012) Extracellular production of Streptomyces exfoliatus poly(3-hydroxybutyrate) depolymerase in Rhodococcus sp. T104: determination of optimal biocatalyst conditions. Appl Microbiol Biotechnol 93: 1975-1988.
- Uefuji M, Kasuya K, Doi Y (1997) Enzymatic degradation of poly[(R)-3-hydroxybutyrate]: secretion and properties of PHB depolymerase from Pseudomonas stutzeri. Polym Degrad Stab 58: 275-281.
- Kita K, Mashiba S, Nagita M, Ishimaru K, Okamoto K, et al. (1997) Cloning of poly(3-hydroxybutyrate) depolymerase from a marine bacterium, Alcaligenes faecalis AE122, and characterization of its gene product. Biochim Biophys Acta 1352: 113-122.
- Tanio T, Fukui T, Shirakura Y, Saito T, Tomita K, et al. (1982) An extracellular poly(3-hydroxybutyrate) depolymerase from Alcaligenes faecalis. Eur J Biochem 124: 71-77.
- Widdick DA, Dilks K, Chandra G, Bottrill A, Naldrett M, et al. (2006) The twin-arginine translocation pathway is a major route of protein export in Streptomyces coelicolor. Proc Natl Acad Sci 103: 17927-17932.
- Hiraishi T, Komiya N, Matsumoto N, Abe H, Fujita M, et al. (2010) Degradation and Adsorption Characteristics of PHB Depolymerase As Revealed by Kinetics of Mutant Enzymes with Amino Acid Substitution in Substrate-Binding Domain. Biomacromolecules 11: 113-119.
- Ohura T, Kasuya KI, Doi Y (1999) Cloning and characterization of the polyhydroxybutyrate depolymerase gene of Pseudomonas stutzeri and analysis of the function of substrate-binding domains. Appl Environ Microbiol 65: 189-197.
- Hisano T, Kasuya K, Tezuka Y, Ishii N, Kobayashi T, et al. (2006) The crystal structure of polyhydroxybutyrate depolymerase from Penicillium funiculosum provides insights into the recognition and degradation of biopolyesters. J Molec Biol 56: 993-1004.
- Kasuya K, Tezuka Y, Ishii N, Yamagata Y, Shiraki M, et al. (2007) Molecular characterization of the poly(3-hydroxybutyrate) depolymerase gene from Penicillium funiculosum. Macromol Symp 249-250: 540-544.
- Handrick R, Reinhardt S, Focaretes ML, Scandola M, Adamus G, et al. (2001) A new type of thermoalkalophilic hydrolase of Paucimonas lemoignei with high specificity for amorphous polyesters of short chain-length hydroxyalkanoic acids. J Biol Chem 276: 36215-36224.
- Jendrossek D, Hermawan S, Subedi B, Papageorgiou A (2013) Biochemical analysis and structure determination of Paucimonas lemoignei poly(3-hydroxybutyrate) (PHB) depolymerase PhaZ7 muteins reveal the PHB binding site and details of substrate-enzyme interactions. Mol Microbiol 90: 649-664.
Citation: Blevins HM, Blue MD, Cobbs BD, Ricotilli TA, Kyler SL, et al. (2018) Characterization of an Extracellular Polyhydroxyalkanoate Depolymerase from Streptomyces sp. SFB5A. J Bioremediat Biodegrad 9: 452. DOI: 10.4172/2155-6199.1000452
Copyright: © 2018 Blevins HM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 6672
- [From(publication date): 0-2018 - Oct 23, 2025]
- Breakdown by view type
- HTML page views: 5691
- PDF downloads: 981