Research Article Open Access
Characterization and QTL Analysis of Oryza longistaminata Introgression Line, pLIA-1, derived from a Cross between Oryza longistaminata and Oryza sativa (Taichung 65) under Non-fertilized Conditions
Emily Gichuhi1, Eiko Himi2, Hidekazu Takahashi3 and Masahiko Maekawa2*
1Graduate School of Environmental and Life Sciences, Okayama University, Japan
2Institute of Plant Science and Resources, Okayama University, Japan
3Bioresource Sciences, Akita Prefectural University, Japan
- Corresponding Author:
- Masahiko Maekawa
Institute of Plant Science and Resources, Okayama University, Japan
Tel: +81864341214
Fax: +81864341249
E-mail: mmaekawa@rib.okayama-u.ac.jp
Received Date: June 16, 2016; Accepted Date: August 18, 2016; Published Date: August 30, 2016
Citation: Gichuhi E, Himi E, Takahashi H, Maekawa M (2016) Characterization and QTL Analysis of Oryza longistaminata Introgression Line, pLIA-1, derived from a Cross between Oryza longistaminata and Oryza sativa (Taichung 65) under Non-fertilized Conditions. J Rice Res 4:174. doi: 10.4172/2375-4338.1000174
Copyright: © 2016 Gichuhi E, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Rice Research: Open Access
Abstract
To meet and sustain the food demands of an ever-increasing world population, improving the yield of major cereal crops such as rice is necessary with sustainable cultivation harmonized with the environment. It is useful to utilize wild rice species as reservoirs of novel traits for breeding low-input adaptable (LIA) crops. Oryza longistaminata, a wild species of rice native only to Africa, possesses the vigorous biomass needed under low-input conditions. Thus, a potential LIA (pLIA) candidate, pLIA-1, showing large biomass, tall culm, large panicle with many primary and secondary branches and thick culms was selected from a selfed progeny of the cross between O. longistaminata and Taichung 65 (T-65), a japonica variety, under non-fertilized conditions. The pLIA-1 performance was superior to that of Koshihikari, Norin 18, T-65 and Nipponbare under fertilized and non-fertilized conditions suggesting that pLIA-1’s characteristics might be useful for breeding low-input adaptable varieties. QTL analysis in F2 of the cross between pLIA-1 and Norin 18 detected 31 QTLs for yield-elated traits under non-fertilized conditions. The pLIA-1 allele had a positive contribution in 20 of the QTLs detected. Importantly, many of the QTLs were identified around regions where O. longistaminata chromosome segments were introgressed into pLIA-1. These results suggests that the QTLs detected in the F2 are important to improve modern varieties for adaptability to low-input conditions.
Keywords
Low-input adaptable; Oryza longistaminata; Wild rice; Oryza sativa
Introduction
In the near future, food insecurity will be a serious global problem, due to an explosively growing population that is predicted to reach 9 billion by 2050 [1]. Therefore, increasing crop productivity of major cereal crops such as rice; to meet the rising demand of an increasing population is mandatory. Rice is a staple food and it accounts for more than 21% of the calorific needs of the world’s population [2].
A major milestone during the green revolution was the development and extensive adoption of semi-dwarf rice cultivars that almost tripled worldwide rice production and greatly enhanced food security [3]. Their high yields were primarily due to their improved harvest index and responsiveness to high inputs, especially nitrogen and water [4]. The extensive usage of semi-dwarf cultivars was, therefore, accompanied by increased inputs of chemical fertilizers and pesticides. This tripled the global grain yield, yet at a high environmental cost. The negative impacts of the current high input agriculture include soil acidification; pollution of rivers, lakes and ground water and the emission of greenhouse gases that may influence global warming [5].
To secure enough food without environmental degradation and excess dependency on inputs, it is necessary to breed new varieties with comparatively high yields and adaptability to low-input conditions. This can be achieved by maximizing crops’ ability to produce biomass and tolerance of various abiotic and biotic stresses through the utilization of genetic resources and distantly related wild relatives. Wild species are reservoirs of latent useful traits for domesticated rice improvement [6]. Although many desirable alleles in wild relatives have, however, not yet been fully exploited, several QTLs for yield-related traits have been identified from Oryza rufipogon [6,7] and Oryza glumaepatula [8]. Of the wild species carrying the AA genome in rice, Oryza longistaminata, native only in Africa, is most distantly related to Oryza sativa [9]. It is a perennial species with resistance to bacterial leaf blight [10-13] and is characterized by strong rhizomes, long anthers and allogamy which are important traits for hybrid seed production [14]. Further, it shows a vigorous biomass under low-input conditions [15,16]. The large biomass of O. longistaminata is considered to be an important trait for breeding low-input adaptable rice. However, its utilization in breeding programs for rice improvement has been very limited, due to developed crossing barriers and hybrid sterility [15,17-19].
We successfully crossed O. longistaminata, with Taichung 65 (T- 65), an O. sativa, and successive self-fertilized plants of the progeny were selected based on large biomass production at a non-fertilized paddy field and called potential low-input adaptable (pLIA) rice line. In this study, we report the characterization of the introgression line, pLIA-1, carrying chromosome segments of O. longistaminata and QTLs for some important yield-related traits detected in F2 of the cross between pLIA-1 and Norin 18, a japonica variety, under non-fertilized conditions.
Materials and Methods
Breeding of the introgression line
Oryza longistaminata, locally known as Mpunga wa Majani (MwM), collected in a valley 50 km away from Mombasa, Kenya, was crossed with T-65, a japonica variety, as a female parent. A single unmatured crossed seed was then subjected to half-strength MS medium [20] culture. F2 population of 169 plants was grown in a greenhouse at the Institute of Plant Science and Resources (IPSR), Okayama University, Kurashiki, Japan. The progeny from the F3 generation was then grown at a non-fertilized paddy field that had been kept without any fertilizer application for more than 20 years at IPSR. Plants showing a large biomass were selected at the F3 generation at the non-fertilized paddy field. Twelve vigorous plants selected at F5 generation were continuously self-fertilized every year. At the F11 generation, a line showing a large biomass under non-fertilized conditions was bred and called potential low-input adaptable (pLIA) rice line, pLIA-1.
To evaluate the contribution of introgressed segment of O. longistaminata (MwM) into pLIA-1 to yield-related traits, F2 plants and F3 plants derived from the cross between pLIA-1 and Norin 18 were grown under non-fertilized conditions and used for QTL mapping.
Characterization of pLIA-1 and its progeny
The agronomic performance of pLIA-1, under both non-fertilized and fertilized conditions, was evaluated with two replicates and compared to T-65, Norin 18, Nipponbare and Koshihikari in 2012 and T-65 and Norin 18 in 2013. In both experiments, 3-week-old seedlings were transplanted with a spacing of 40 cm between rows and 15 cm between plants. Fifty kg/ha of N, P and K were supplied in fertilized field as basal dressing. The following agronomic traits were measured: the number of panicles per plant, total weight of panicles, total weight of the biomass of the plant shoot, 100-grain weight and grain yield. Culm length, flag leaf length, panicle length, fresh panicle-base diameter, fresh culm-base diameter (culm diameter at 5 cm above ground), number of primary branches per panicle, number of secondary branches per panicle, number of spikelets per panicle and spikelet fertility were measured on the tallest culm of a given plant. Grain length and grain width were measured in 30 grains with 3 replicates. The rough harvest index was calculated as the panicle weight/total plant weight in 2012. Days to heading were from sowing date to the emergence of the first panicle.
F2 and F3 plants derived from the cross between pLIA-1 and Norin 18 were only grown under non-fertilized conditions in 2011 and 2012, respectively. A total of 14 yield-related traits were measured and evaluated.
Analysis of variance (ANOVA) was performed using R software. The Tukey’s test (P=0.05) was used to test the differences between means, where significant differences were detected by ANOVA.
Graphical genotyping of pLIA-1
Leaf samples from seedlings of MwM, Norin 18, T-65 and pLIA- 1 were collected and dried at 50°C overnight. Genomic DNA was extracted from the leaf tissues using a modified procedure as described by Kawasaki [21]. PCR reaction was prepared by mixing 3.5 μl of distilled water, 20 pmol of 0.5 μl forward primer, 20 pmol of 0.5 μl reverse primer, 5 μl of Quick Taq (TOYOBO, Japan) and 5ng of the extracted DNA and 5 ng of the extracted DNA. Amplification was performed as follows: the initial denaturing step at 95°C for 7 min, 30 cycles for 45 sec at 95°C, 30 sec at 55°C and 30 sec at 72°C. Electrophoresis was done in a 3% agarose gel for 90 minutes. The band pattern of the samples was observed in UV-lighting after staining with Ethidium bromide.
QTL analysis
The F2 plants derived from the cross between pLIA-1 and Norin 18 were genotyped using 35 SSR markers found to be polymorphic between pLIA-1 and Norin 18, out of 111 SSR markers genome-widely distributed. To validate the QTLs identified in the F2 population, 230 F3 plants were genotyped using additional 8 SSR markers in the vicinity of the QTL identified on the distal end of the long arm of chromosome 8. The genetic linkage map was constructed using Mapmaker/Exp version 3.0. Composite interval mapping was performed for QTL analysis using the software Window QTL Cartographer 2.5 [22]. For each of the traits, the LOD threshold was determined at significant probability level of 5% by computing 1000 permutations, in both populations.
Results and Discussion
The breeding process
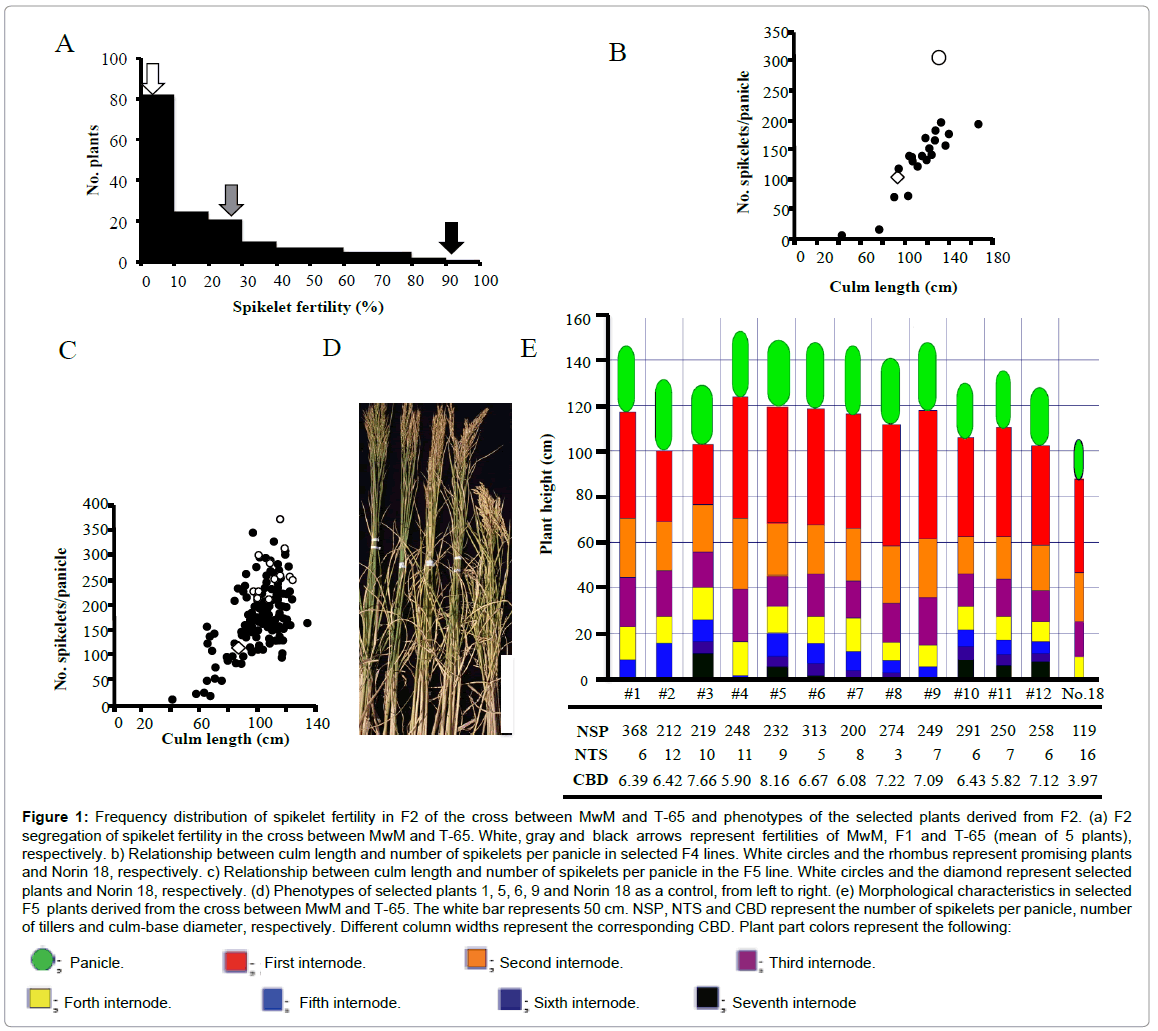
Crossing barriers and hybrid sterility highly developed between O. longistaminata and O. sativa [17] hindered the direct analysis of some important traits in the F2 population. Therefore, many researchers bred backcrossed populations for genetic analyses. Maekawa et al. [23] reported that O. longistaminata from Ethiopia has comparatively good crossing ability with O. sativa and an F2 population could be produced. Iwamoto et al. [24] demonstrated that O. longistaminata from Kenya is closely related to that from Ethiopia, based on the Catalase gene structure. In this study, O. longistaminata from Kenya was successfully crossed with T-65 as a pollen-donor parent and an F2 population was obtained. A single F1 plant of the cross between MwM and T-65 showed low spikelet fertility of 27%. Although 107 of the 169 plants showed extremely high sterility in the F2, a few highly fertile plants segregated, as shown in Figure 1A. In order to obtain large biomass plants derived from the cross, descendants from fertile F2 plants were grown in the non-fertilized field. Thus, one promising plant was selected based on its large biomass production and large panicle. The relationship between the culm length and the number of spikelets per panicle of F4 plants selected from the F3 plant demonstrated that plants carrying large numbers of spikelets per panicle had relatively long culm lengths (Figure 1B). Furthermore, the segregation of F5 plants was examined and the segregation pattern was found to be similar to that of F4 plants (Figure 1C). Twelve plants (#1 to #12) showing large numbers of spikelets per panicle were, therefore, selected. These selected plants were characterized by large panicles, large numbers of spikelets per panicle, long culms, many elongated internodes, thick culm base diameters and few tillers, compared with those of Norin 18 (Figures 1D and 1E). Although plants carrying large panicles and long culms tend to lodge easily, these selected plants were highly tolerant to lodging due to the very thick culm-base diameter. Of the 12 selected plants, #1 plant with more spikelets per panicle was selectively grown for further self-fertilization at the non-fertilized field. After 11 generations of selffertilization, this line was named potential low-input adaptable (pLIA) line, pLIA-1.
Figure 1: Frequency distribution of spikelet fertility in F2 of the cross between MwM and T-65 and phenotypes of the selected plants derived from F2. (a) F2 segregation of spikelet fertility in the cross between MwM and T-65. White, gray and black arrows represent fertilities of MwM, F1 and T-65 (mean of 5 plants), respectively. b) Relationship between culm length and number of spikelets per panicle in selected F4 lines. White circles and the rhombus represent promising plants and Norin 18, respectively. c) Relationship between culm length and number of spikelets per panicle in the F5 line. White circles and the diamond represent selected plants and Norin 18, respectively. (d) Phenotypes of selected plants 1, 5, 6, 9 and Norin 18 as a control, from left to right. (e) Morphological characteristics in selected F5 plants derived from the cross between MwM and T-65. The white bar represents 50 cm. NSP, NTS and CBD represent the number of spikelets per panicle, number of tillers and culm-base diameter, respectively. Different column widths represent the corresponding CBD. Plant part colors represent the following:

Characterization of the pLIA-1
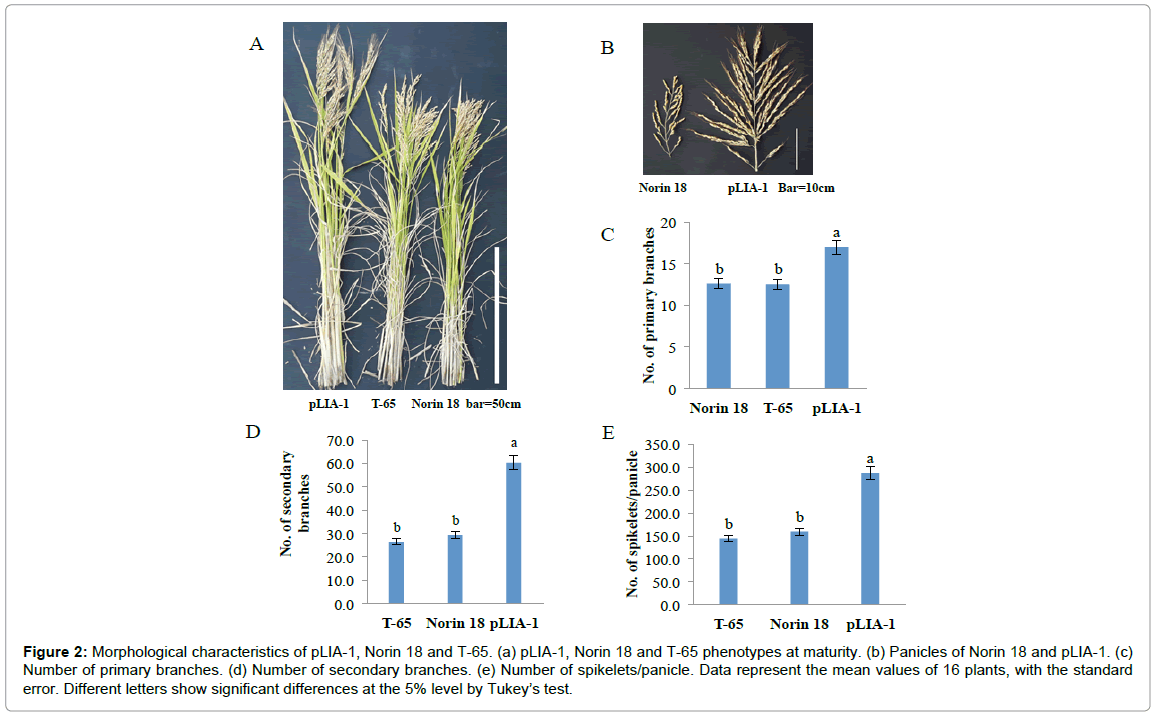
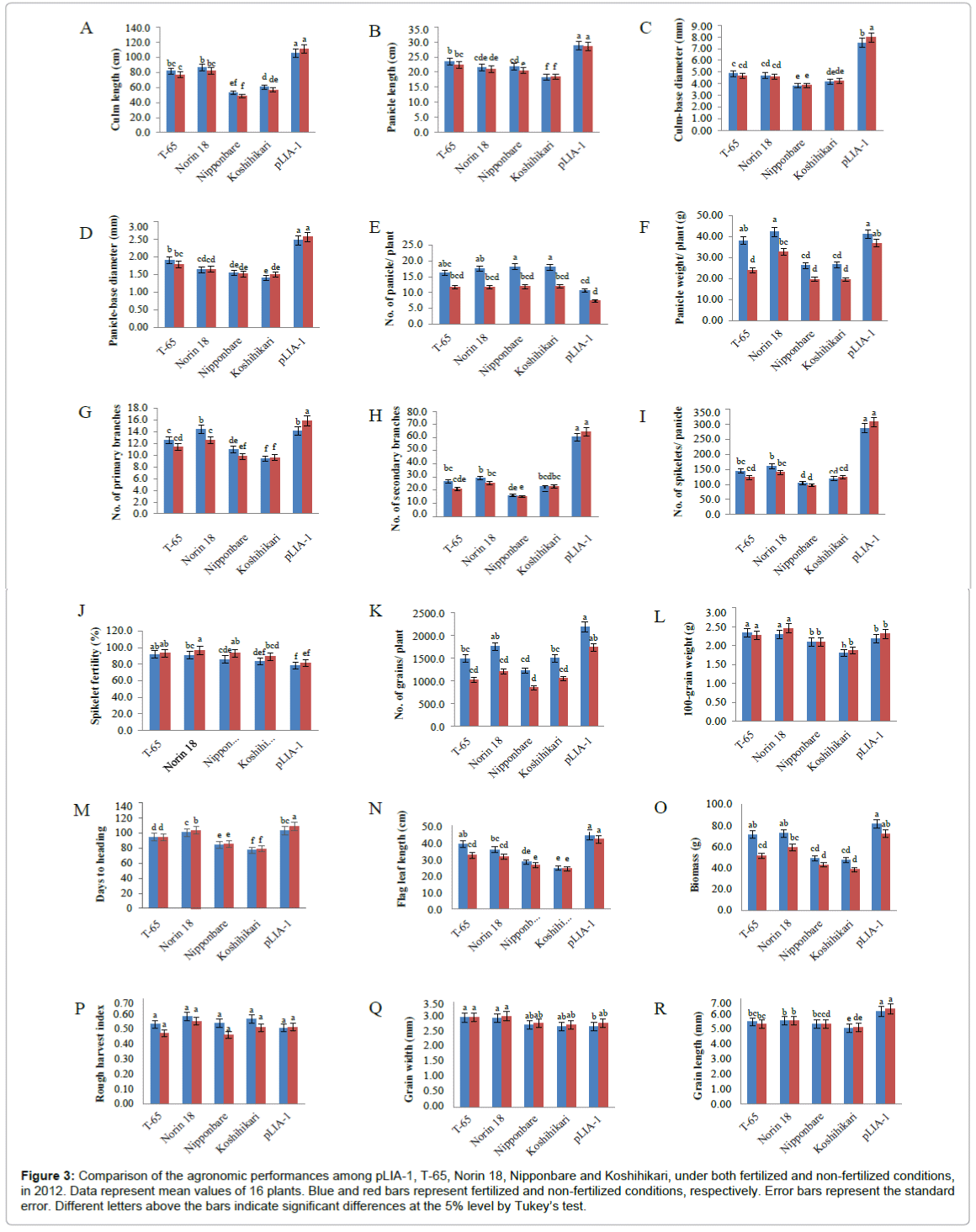
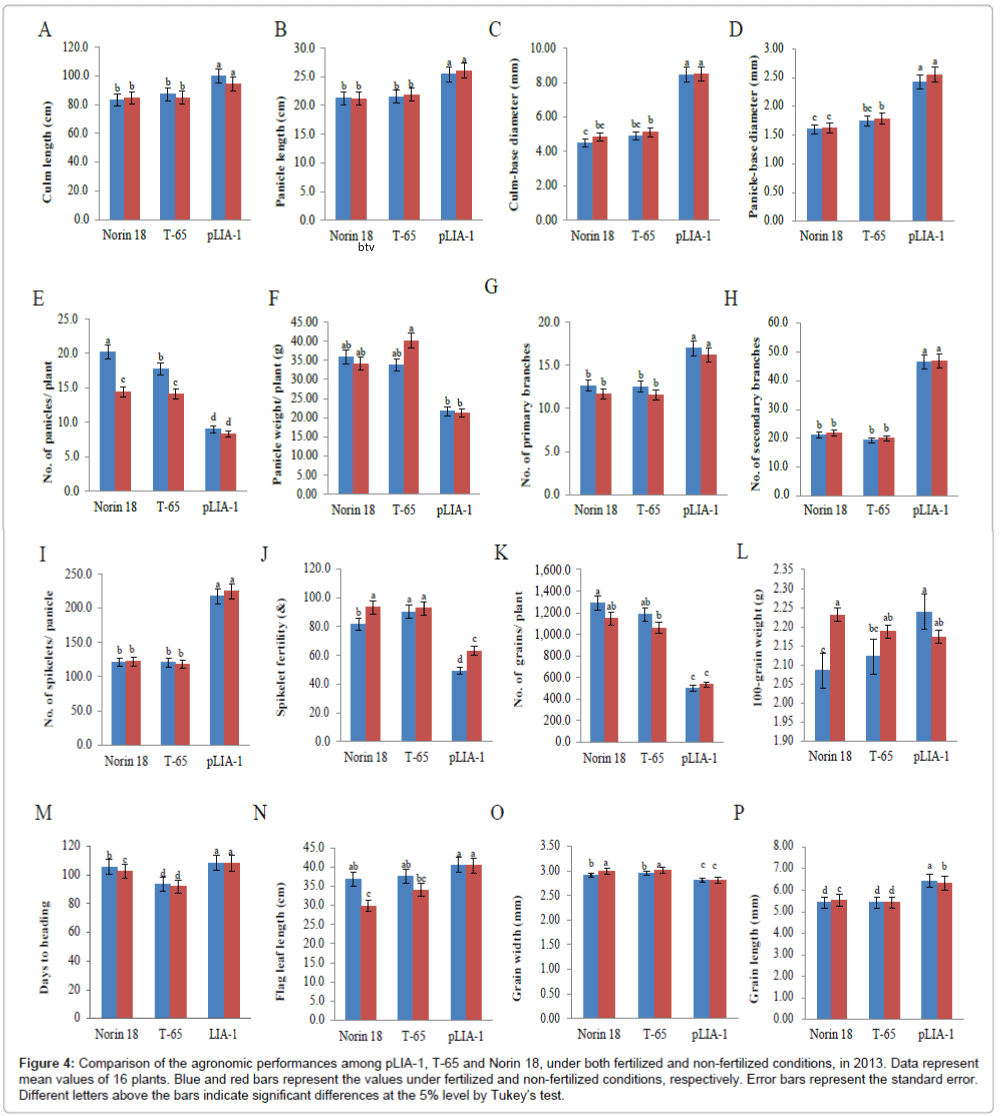
Recently, it has been reported that most of the semi-dwarf rice varieties have high numbers of unproductive tillers, small panicles and are susceptible to lodging in directly seeded conditions [25]. These traits are identified as the major constraints on improving yields in these varieties; whose yield potential has stagnated due to their plant type. Therefore, the concept of ideal plant architecture (IPA) has been proposed and demonstrated using plants with low tiller numbers with few unproductive tillers, more grains per panicle than in the currently cultivated varieties and thick and sturdy stems [26,27]. As shown in Figure 2, pLIA-1 had a very large number of spikelets per panicle with a large number of primary and secondary branches, compared with T-65 and Norin 18 (Figures 2A and 2B). Significantly larger numbers of primary and secondary branches per panicle resulted in a larger number of spikelets per panicle (Figures 2C-2E). Therefore, the agronomic traits of pLIA-1 were compared to those of T-65, Nipponbare and Koshihikari together with Norin 18 in 2012 and Norin 18 and T-65 in 2013, under both fertilized and non-fertilized conditions (Figures 3 and 4). Significant differences in the culm length, panicle length, culm-base diameter, panicle-base diameter, and number of secondary branches, number of spikelets per panicle and grain length between pLIA-1 and other varieties were observed under both conditions, in both years (Figures 3A-3D, 3H, 3I, 3R, 4A-4D, 4H, 4I and 4P). T-65 showed a significant decrease in panicle weight, total grain yield, flag leaf length and biomass in 2012 (Figures 3F, 3K, 3N and 3O) and number of panicles in 2013 (Figure 4E) under non-fertilized conditions. On the other hand, Norin 18 showed significantly decreased panicle weight, number of primary branches and number of grains/plant in 2012 (Figures 3F, 3G and 3K) and number of panicles, days to heading and flag leaf length under non-fertilized conditions in 2013 (Figures 4E, 4M and 4N). Significant increases in the culm-base diameter, number of primary branches and days to heading were observed under nonfertilized conditions for pLIA-1 in 2012 (Figures 3C, 3G and 3M). Increases in the panicle-base diameter, number of secondary branches and number of spikelets per panicle were also observed under nonfertilized conditions in both years; however, the increase was not significant (Figures 3D, 3H, 3I, 3L, 4D, 4H, 4I and 4L). Although pLIA-1 showed significantly shorter grain length under non-fertilized conditions in 2013 (Figure 4P), it did not show a significant reduction in any other trait under non-fertilized conditions. These results suggest that pLIA-1 performance is superior under non-fertilized conditions, compared to other varieties. The characteristics of the pLIA-1 selected under non-fertilized conditions are, hence, comparable to IPA. Overall, in comparison to other varieties, pLIA-1 was significantly superior in most of the traits measured. However, the spikelet fertility of pLIA-1 was found to be significantly lower than in other varieties. The spikelet fertility recorded in 2013 was markedly low and may have had a direct effect on the total grain yield. The inconsistence observed in spikelet fertility and a few other traits between 2012 and 2013 may be due to environmental effect.
Figure 2: Morphological characteristics of pLIA-1, Norin 18 and T-65. (a) pLIA-1, Norin 18 and T-65 phenotypes at maturity. (b) Panicles of Norin 18 and pLIA-1. (c) Number of primary branches. (d) Number of secondary branches. (e) Number of spikelets/panicle. Data represent the mean values of 16 plants, with the standard error. Different letters show significant differences at the 5% level by Tukey’s test.
Figure 3: Comparison of the agronomic performances among pLIA-1, T-65, Norin 18, Nipponbare and Koshihikari, under both fertilized and non-fertilized conditions, in 2012. Data represent mean values of 16 plants. Blue and red bars represent fertilized and non-fertilized conditions, respectively. Error bars represent the standard error. Different letters above the bars indicate significant differences at the 5% level by Tukey’s test.
Figure 4: Comparison of the agronomic performances among pLIA-1, T-65 and Norin 18, under both fertilized and non-fertilized conditions, in 2013. Data represent mean values of 16 plants. Blue and red bars represent the values under fertilized and non-fertilized conditions, respectively. Error bars represent the standard error. Different letters above the bars indicate significant differences at the 5% level by Tukey’s test.
Graphical genotyping of pLIA-1
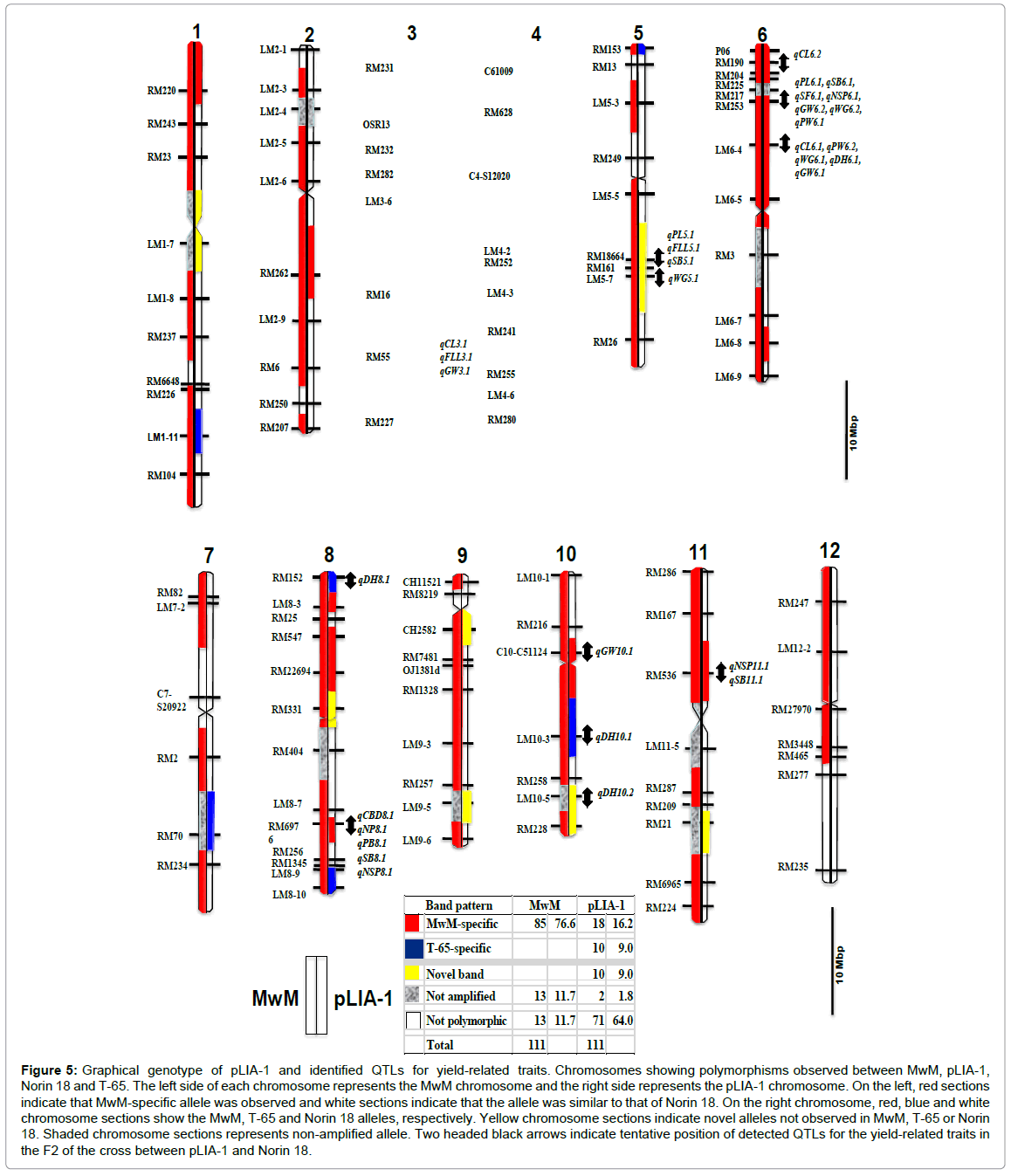
In order to utilize pLIA-1’s characteristics in breeding programs, introgressed segments in pLIA-1 were needed to be revealed. Tian et al. [28] found that QTLs derived from O. rufipogon introgressed segments in a set of 159 introgressed lines were usually associated with improvement of the target trait (panicles per plant, grains per panicle and filled grains per panicle). In this study, pLIA-1 was found to possess introgressed chromosome segments of O. longistaminata and T-65 by using genome-wide SSR markers. As shown in Figure 5, MwM showed specific polymorphisms in 85 (76.6% frequency) of 111 markers used. Thirteen markers were not amplified in MwM. These non-amplified markers were distributed in 10 chromosomes, except chromosomes 5 and 12 in MwM. Since O. longistaminata is predicted to carry a highly rearranged DNA sequence (Maekawa, unpublished), it is likely that 13 of the markers were not amplified. Additionally, other 13 markers were not polymorphic among the markers used. However, it was observed that pLIA-1 showed MwM-specific band patterns in 18 of 85 markers in 7 chromosomes, except chromosomes 4, 5, 7, 9 and 12 (Figure 5). Ten markers showed T-65-specific band patterns on chromosomes 1, 4, 5, 7, 8 and 10. Consequently, pLIA-1 was found to carry 20 MwM-specific markers including 2 non-amplified markers on chromosomes 1, 2, 3, 6, 8, 10 and 11 with a frequency of 18.0%. In particular, the short arm of chromosome 6 of O. longistaminata is presumed to be introduced into pLIA-1. Relatively few segments of O. longistaminata were introgressed into pLIA-1 chromosomes. Of 111 genome-1 wide SSR markers used, only 18 O. longistaminata specific markers were observed with a frequency of 16.2%. However, pLIA-1 was shown to exhibit the large biomass characteristic under non-fertilized conditions. This suggests that the large biomass and lowinput tolerance characteristics may be controlled by small segments of the O. longistaminata chromosome segments. On the other hand, novel introgressed chromosome segments observed on chromosomes 1, 5, 8, 9, 10 and 11 may have been caused by changed short sequence repeat numbers through successive self-fertilization or more likely outcrossing during the early stages of the breeding process of pLIA-1.
Figure 5: Graphical genotype of pLIA-1 and identified QTLs for yield-related traits. Chromosomes showing polymorphisms observed between MwM, pLIA-1, Norin 18 and T-65. The left side of each chromosome represents the MwM chromosome and the right side represents the pLIA-1 chromosome. On the left, red sections indicate that MwM-specific allele was observed and white sections indicate that the allele was similar to that of Norin 18. On the right chromosome, red, blue and white chromosome sections show the MwM, T-65 and Norin 18 alleles, respectively. Yellow chromosome sections indicate novel alleles not observed in MwM, T-65 or Norin 18. Shaded chromosome sections represents non-amplified allele. Two headed black arrows indicate tentative position of detected QTLs for the yield-related traits in the F2 of the cross between pLIA-1 and Norin 18.
Correlations between yield-related traits
To reveal the important QTLs for yield-related traits, segregation patterns of the traits and correlations between them were examined in F2 of the cross between pLIA-1 and Norin 18. In most of the traits measured, segregation patterns of normal distribution were observed (Supplementary Figure 1) and transgressive segregations were found in all the traits. In order to understand QTL cluster, correlations among agronomic traits were examined in the F2. It was found that culmbase diameter was significantly positively correlated with panicle traits (panicle length, panicle weight, number of primary branches, number of secondary branches and number of spikelets per panicle) and flag leaf length (Table 1). The panicle traits were further observed to be positively correlated to each other. The number of spikelets per panicle was strongly correlated to the number of secondary branches (Table 1). Further, significant positive correlations between flag leaf length and number of primary branches, number of secondary branches and number of spikelets per panicle were observed (Table 1).
| Trait | Culm length (cm) | Panicle length (cm) | No. of panicles | Culm-basediamter (mm) | Flag leaf length (cm) | One panicle weight (g) | No. of primary branches | No. of secondary branches | No. of secondary spikelets/panicle | Spikelet fertility (%) | 100-grain weight (g) | Grain length (mm) | Grain width (mm) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Panicle length (cm) | -0.118 | ||||||||||||
| No. of panicles | 0.231** | 0.137 | |||||||||||
| Culm-basediamter (mm) | 0.153 | 0.237** | 0.058 | ||||||||||
| Flag leaf length (cm) | -0.307** | 0.407** | -0.003 | 0.225** | |||||||||
| One panicle weight (g) | 0.095 | 0.19* | 0.021 | 0.34** | 0.154 | ||||||||
| No. of primary branches | 0.043 | 0.124 | 0.007 | 0.477** | 0.248** | 0.265** | |||||||
| No of secondary branches | 0.081 | 0.376** | 0.167* | 0.508** | 0.368** | 0.499** | 0.379** | ||||||
| No. of spikelets/ panicle | 0.072 | 0.319** | 0.173* | 0.521** | 0.349** | 0.494** | 0.534** | 0.847** | |||||
| Spikelet fertility (%) | 0.113 | 0.068 | 0.064 | -0.02 | -0.105 | 0.546** | 0.002 | 0.079 | 0.041 | ||||
| 100-grain weight (g) | -0.218** | 0.006 | -0.019 | -0.032 | 0.052 | 0.004 | -0.027 | -0.163* | -0.097 | -0.275** | |||
| Grain length (mm) | -0.155* | 0.125 | -0.051 | 0.077 | 0.104 | 0.114 | 0.021 | 0.039 | -0.025 | -0.108 | 0.583** | ||
| Grain width (mm) | -0.117 | -0.106 | -0.06 | 0.046 | -0.057 | -0.098 | 0.056 | -0.123 | 0.04 | -0.317** | 0.47** | -0.165* | |
| Days to heading | 0.063 | -0.171* | -0.151 | -0.148 | -0.121 | -0.25** | -0.184* | -0.169* | -0.220** | -0.267** | -0.024 | 0.088 | -0.066 |
Table 1: Correlation coefficient of yield-related traits in F2 of the cross between pLIA-1 and Norin 18 under non-fertilized conditions.
QTL analysis
To explore the genetic resources from wild rice, several populations derived from crosses between various cultivars and wild rice have been used for identification of important QTLs for agronomic traits [7,8,25]. In particular, Xiao et al. [6] detected a total of 68 QTLs for 12 traits using a backcross population derived from a wild rice (O. rufipogon) and cultivated rice. In this study, 31 QTLs for 13 yield-related traits were detected on chromosomes 3, 5 6, 8, 10 and 11 in the F2 of the cross between pLIA-1 and Norin 18 (Table 2 and Figure 5). These included 3 QTLs for culm length, 2 QTLs for panicle length, 1 QTL for number of panicles per plant, 1 QTL for culm-base diameter, 2 QTLs for flag leaf length, 2 QTLs for panicle weight, 1 QTL for number of primary branches per panicle, 4 QTLs for number of secondary branches per panicle, 3 QTLs for number of spikelets per panicle, 1 QTL for spikelet fertility, 3 QTLs for 100 grain weight, 4 QTLs for days to heading and 4 QTLs for grain width (Table 2). The QTLs were distributed on chromosomes 3, 5, 6, 8, 10 and 11(Figure 5). In 20 of the QTLs identified, pLIA-1 had a positive contribution to the trait. QTLs for strongly correlated traits were observed to be localized near the same region on the chromosome. These clusters of QTLs were observed on chromosomes 3, 5, 6 and 8 (Figure 5). Further analysis using an F3 population revealed that the region around RM6976 on chromosome 8 carried a crucial QTL cluster for culm-base diameter, panicle-base diameter and number of primary branches (Figure 5 and Table 3). This is considered to be caused by a strong positive correlation observed between the number of primary branches, number of secondary branches and number of spikelets per panicle. Hence, it is plausible that QTLs for number of primary branches and number of spikelets per panicle were identified in the same QTL cluster in the F2 population. These results strongly suggest that O. longistaminata has great potential for utilization in yield improvement and especially under low-input conditions despite the low spikelet fertility. Among the traits transferred in interspecific crosses using wild rice relatives, spikelet sterility is especially a serious constraint.
| Trait | QTL | Chr. | Marker | LOD | Additive effect | r2 |
|---|---|---|---|---|---|---|
| Culm length | qCL3.1 | 3 | RM55 | 3.4 | -3.69 | 0.07 |
| qCL6.2 | 6 | LM6_4 | 6.4 | 6.45 | 0.16 | |
| qCL6.1 | 6 | RM190 | 6.3 | 5.75 | 0.12 | |
| Panicle length | qPL5.1 | 5 | RM18664 | 5.9 | 1.24 | 0.25 |
| qPL6.1 | 6 | RM253 | 6.7 | -1.29 | 0.18 | |
| No. of panicles/ plant | qNP8.1 | 8 | RM6976 | 4.6 | -1.35 | 0.11 |
| Culm-base diameter | qCBD8.1 | 8 | RM6976 | 7.1 | 0.44 | 0.15 |
| Flag leaf length | qFLL3.1 | 3 | RM55 | 3.2 | 2.83 | 0.07 |
| qFLL5.1 | 5 | RM18664 | 3.0 | 2.42 | 0.14 | |
| Panicle weight | qPW6.1 | 6 | RM253 | 9.3 | -0.23 | 0.29 |
| qPW6.2 | 6 | LM6_4 | 3.7 | -0.24 | 0.08 | |
| No. of primarybranches | qPB8.1 | 8 | RM6976 | 15.3 | 1.74 | 0.29 |
| No. of secondary | qSB5.1 | 5 | RM18664 | 5.1 | 2.67 | 0.18 |
| branches | qSB6.1 | 6 | RM253 | 4.9 | -4.48 | 0.15 |
| qSB8.1 | 8 | RM6976 | 3.2 | 2.63 | 0.06 | |
| qSB11.1 | 11 | RM536 | 3.5 | 3.77 | 0.07 | |
| No. of spikelets/ | qNSP6.1 | 6 | RM253 | 3.2 | -12.77 | 0.10 |
| panicle | qNSP8.1 | 8 | RM6976 | 6.0 | 17.57 | 0.12 |
| qNSP11.1 | 11 | RM536 | 3.0 | 13.55 | 0.06 | |
| Spikelet fertility | qSF6.1 | 6 | RM253 | 30.9 | -1.57 | 0.60 |
| Weight of 100-grains | qWG5.1 | 5 | LM5_7 | 3.7 | -0.16 | 0.39 |
| qWG6.2 | 6 | LM6_4 | 9.1 | -0.11 | 0.23 | |
| qWG6.1 | 6 | RM253 | 7.9 | 0.00 | 0.16 | |
| Days to heading | qDH6.1 | 6 | LM6_4 | 24.2 | 2.95 | 0.33 |
| qDH8.1 | 8 | RM152 | 8.7 | -1.92 | 0.14 | |
| qDH10.1 | 10 | LM10_3 | 10.6 | 2.17 | 0.22 | |
| qDH10.2 | 10 | LM10_5 | 3.5 | 0.94 | 0.04 | |
| Grain width | qGW3.1 | 3 | RM55 | 13.7 | -0.06 | 0.23 |
| qGW6.2 | 6 | LM6_4 | 7.5 | -0.05 | 0.12 | |
| qGW6.1 | 6 | RM253 | 5.5 | 0.00 | 0.07 | |
| qGW10.1 | 10 | C51124 | 3.9 | 0.03 | 0.06 |
Table 2: Marker position, LOD score, additive effect and contribution rate of QTL for yield-related traits identified in the F2 population.
| Trait | QTL | Marker | LOD | Additive effect | r2 |
|---|---|---|---|---|---|
| Culm-base diameter | qCBD8.1 | RM6976 | 8.8 | 0.39 | 0.16 |
| Panicle-base diameter | qPBD8.1 | RM210 | 2.2 | 0.06 | 0.04 |
| No. of primary branches | qPB8.1 | RM6976 | 18.2 | 1.16 | 0.30 |
| RM210 | 17.1 | 1.12 | 0.29 |
Table 3: QTL for yield-related traits validated in the F3 population on chromosome 8 QTL cluster region.
A QTL for spikelet fertility was identified on chromosome 6 with a very high LOD score of 30.9 and 60% contribution to the total phenotypic variation (Table 2), suggesting that this QTL might be the major cause of the low spikelet fertility of pLIA-1. In fact, this QTL was located near the same chromosome region where a QTL for pollen and spikelet fertility was previously identified in a cross using O. longistaminata [29].
QTL clusters of functionally related genes are of great interests in crop improvement. A total of 4 QTL clusters on chromosome 3, 5, 6 and 8 were observed (Figure 5). Highly significant correlations were also observed between the yield-related traits whose QTLs’ were observed to cluster in the same chromosome locations (Table 1). In previous QTL analysis it has been observed that QTL for significantly correlated traits usually had same chromosome location [8,28]. The QTL cluster on chromosome 8 was identified near the same chromosome region where the WFP (Wealthy Farmer’s Panicle) was found to be located [30]. This result suggest that these traits are either controlled by strongly linked genes or are as a result of pleiotropism of a single gene locus located at the regions where QTL clusters were observed. Previously, Ookawa et al. [31] reported that the APO1 gene of Habataki on chromosome 6 increased spikelet number together with thicker culm through increased size of inflorescence meristem, hence, the higher spikelet number induced culm thickness pleiotropically. The WFP gene of ST-12 which encodes OsSPL14 (Squamosa Promoter Binding Protein-Like14) drastically increases primary branch number, resulting in increased number of spikelets per panicle [30]. Since in the report, inflorescence meristem of ST-12 was found to be larger than that of Nipponbare, ST-12 was presumed to have thick culms. Thus, it is plausible that the QTL of culm-base diameter was located near the QTL for primary branch number on chromosome 8 based on significantly positive correlation between culm-base diameter and panicle traits observed in this study. Further, there was no correlation between culmbase diameter and culm length in this study suggesting that different culm length plants could be bred with thick culm and larger panicles. Taken together, O. longistaminata is therefore suggested to carry several useful traits under low-input conditions.
The pLIA-1 line reported here is considered to have high potential for low-input adaptability. Identification of QTLs for yield-related traits under non-fertilized conditions further proves its potential for utilization in yield improvement of rice. This line could therefore be utilized to introgress high productivity under low-input conditions to elite rice varieties. Improvement of nitrogen use efficiency has been proposed as a target for the Second Green Revolution [32]. Hence, “low-input and high output” agriculture is required for sustainability. Further analysis of this line and identification of genes governing the QTLs of agronomic importance is necessary for better understanding of its tolerance to low-input conditions.
Conclusion
In order to utilize O. longistaminata as gene resources, some selected plants which showed vigorous biomass under non-fertilized conditions were developed from the F2 of the cross between MwM, O. longistaminata collected in Kenya and T-65, O. sativa. Out of the selected plants, pLIA-1 showed tolerance to non-fertilized conditions compared to the other varieties, hence was named potential Low-input Adaptable-1. The pLIA-1 was subjected to polymorphic analysis against Norin 18 using SSR markers. Although pLIA-1 carried 20 MwMspecific segments, very important QTLs for panicle-related traits were especially found to be intensively located on the distal region of long arm of chromosome 8 in F2 of the cross between pLIA-1 and Norin 18.
Acknowledgements
This research was funded by the Japan Science and Technology Agency (JST)/Japan International Cooperation Agency (JICA), the Science and Technology Research Partnership for Sustainable Development (SATREPS) and the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
References
- Alexandratos N, Bruinsma J (2012) World agriculture towards 2030/2050: The 2012 revision. ESA Working Paper No. 12-03, Rome.
- Fitzgerald MA, McCouch SR, Hall RD (2009) Not just a grain of rice: the quest for quality. Trends Plant Sci 14: 133-139.
- Tilman D (1998) The greening of the green revolution. Science 396: 211-212.
- Matson PA, Parton WJ, Power AG, Swift MJ (1997) Agricultural intensification and ecosystem properties. Science 277: 504-509.
- Ravishankara AR, Daniel JS, Portmann RW (2009) Nitrous Oxide (N2O): The dominant ozone-depleting substance emitted in the 21st Century. Science 326: 123-125.
- Xiao JH, Li JM, Grandillo S, Ahn SN, Yuan LP, et al. (1998) Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryzarufipogon. Genetics 150: 899-909.
- ReddyCS, BabuAP, SwamyBPM, SarlaN (2007) Insight Into Genes underlying yield enhancing QTLs from O. rufipogon. Rice Genetics Newsletter 23: 53-55.
- Brondani C, Rangel PHN, Brondani RPV, Ferreira ME (2002) QTL mapping and introgression of yield-related traits from Oryzaglumaepatula to cultivated rice (Oryza sativa)using microsatellite markers. TheorAppl Genet 104: 1192-1203.
- Causse MA, Fulton TM, Cho YG, Ahn SN, Chunwongse J, et al. (1994) Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138: 1251-1274.
- KhushGS,BacalangoE,OgawaT(1990)ANewGeneforResistanceto Bacterial Blight from O. longistaminata. Rice Genetics Newsletter 7: 121-122.
- Khush GS, Mackill DJ, Sidhu GS (1989) Breeding rice for resistance to bacterial blight. In: Bacterial blight of rice. Proceeding of international workshop on bacterial blight of rice, IRRI, Manila, Philippines.pp: 207-217.
- Ronald PC, Albano B, Tabien R, Abenes L, Wu KS, et al. (1992) Genetic and physical analysis of the rice bacterial-blight disease resistance locus, Xa21. Molecular & General Genetics 236: 113-120.
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, et al. (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804-1806.
- Virmani SS, Aquino RC, Khush GS (1982) Heterosis breeding in rice (Oryza sativa L). Theoretical and Applied Genetics 63: 373-380.
- Sacks EJ, Roxas JP, Cruz MTS (2003) Developing perennial upland rice II:Field performance of S-1 families from an intermatedOryza sativa/O. longistaminata population. Crop Sci 43: 129-134.
- Yang H, Hu L, Hurek T, Reinhold-Hurek B (2010) Global characterization ofthe root transcriptome of a wild species of rice, Oryzalongistaminata, by deepsequencing. BMC Genomics 11: 705.
- Chu YE, Oka HI (1970) The genetic basis of crossing barriers between Oryza Perennis Subsp. barthii and its related taxa. Evolution 24:135-144.
- Hu F, Wang D, Zhao X, Zhang T, Sun H, et al. (2011) Identification of rhizome-specific genes by genome-wide differential expression analysis in Oryzalongistaminata. BMC Plant Biol 11:18-31.
- Zong Y, Huang L, Zhang T, Qin Q, Wang W, et al. (2014) DifferentialmicroRNA expression between shoots and rhizomes in Oryzalongistaminatausing high-throughput RNA sequencing. The Crop J 2: 102-109.
- Murashige T, Skoog FK (1962) A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiologia Plantarum15:473-497.
- Kawasaki T (1997) In: Shimamoto K, Sasaki T (ed.) Simplified extraction method of rice genomic DNA for PCRanalysis. PCR-based Experimental Protocol in Plants, Shujyunsya, Tokyo. pp: 67-68.
- Wang S, Basten CJ, Zeng ZB (2007) Windows QTL Cartographer 2.5.Department of Statistics, North Carolina State University, Raleigh, USA.
- MaekawaM, RikiishiK, MatsuuraK, NodaK (1996) Genetic analysis of rhizomatous trait of wild species (Oryzalongistaminata) in rice (in Japanese).Breed Sci 46: 323
- Iwamoto M, MaekawaM, SaitoA, HigoH, HigoK (1998) Evolutionary relationship of plant catalase genes inferred from exon âÂ?Â?intron structures: Isozyme divergence after the separation of monocots and dicots. Appl Genet 97: 9-19.
- Peng S, Khush GS, Virk P, Tang Q, Zou Y (2008) Progress in ideotype breeding to increase rice yield potential. FieldCropsRes108:32-38.
- Khush GS (1995) Modern varieties: Their real contribution to food supply andequity. Geo J 35: 275-284.
- Jiao Y, Wang Y, Xue D, Wang J, Yan M, et al. (2010) Regulation of OsSPL14by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42: 541-544.
- Tian F, Li DJ, Fu Q, Zhu ZF, Fu YC, et al. (2006) Construction of introgressionlines carrying wild rice (OryzarufipogonGriff.) segments in cultivated rice (Oryza sativaL.) background and characterization of introgressed segments associated with yield-related traits. TheorAppl Genet 112: 570-580.
- Chen Z, Hu F, Xu P, Li J, Deng X, et al. (2009) QTL analysis for hybrid sterilityand plant height ininterspecific populations derived from a wild rice relative, Oryzalongistaminata. Breed Sci 59:441-445.
- MiuraK, IkedaM, MatsubaraA, SongX, ItoM, etal. (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42: 545-549.
- Ookawa T, Hobo T, Yano M, Murata K, Ando T, et al. (2010) New approach for rice improvement using apple tropic QTL gene for lodging resistance and yield. Nat Commun 1: 132.
- d eRibou SDB, Douam F, Hamant O, Frohlich MW, Negrutiu J (2013) Plantscience and agricultural productivity: Why are we hitting the yield ceiling? Plant Sci 210: 159-176.
Relevant Topics
- Basmati Rice
- Drought Tolerence
- Golden Rice
- Leaf Diseases
- Long Grain Rice
- Par Boiled Rice
- Raw Rice
- Rice
- Rice and Aquaculture
- Rice and Nutrition
- Rice Blast
- Rice Bran
- Rice Diseases
- Rice Economics
- Rice Genome
- Rice husk
- Rice production
- Rice research
- Rice Yield
- Sticky Rice
- Stress Resistant Rice
- Unpolished Rice
- White Rice
Recommended Journals
Article Tools
Article Usage
- Total views: 12530
- [From(publication date):
December-2016 - Apr 03, 2025] - Breakdown by view type
- HTML page views : 11582
- PDF downloads : 948