Cellulolytic Bacillus may or may not Produce Îò-Glucosidase due to their Environmental Origin âÃâ¬Ãâ A Case Study
Received: 31-Aug-2017 / Accepted Date: 11-Sep-2017 / Published Date: 14-Sep-2017 DOI: 10.4172/2155-6199.1000412
Abstract
Microbial cellulases have been drawing attention worldwide because of their massive capacity to process the most abundant cellulosic biomass into sustainable biofuels and other valuable products. Profitable biomass conversion processes are highly dependent on the use of efficient enzymes for lignocellulose degradation. Among the cellulose degrading enzymes, β-glucosidases are essential for efficient hydrolysis of cellulosic biomass as they relieve the inhibition of the cellobiohydrolases and endoglucanases by reducing cellobiose accumulation. In this study, cellulolytic bacteria with potential β-glucosidases activity were isolated and screened from biogas plant effluent and dairy effluent near Jahangirnagar University campus. From initial screening, a total of 16 isolates were found to have cellulolytic activity, among them three isolates (B1, B5, D4) were selected based on their superior results. All the three bacterial isolates were identified as B. subtilis (B1), Bacillus amyloliquefaciens (B5) and B. subtilis (D4) respectively based on their morphological, biochemical and molecular characteristics. The β- glucosidases activity of these three potential cellulolytic bacteria was performed by measuring the release of PNP using pNPG as a substrate and interestingly D4 strain was resulted with β-glucosidases negative where B1 strain was found to have efficient for β-glucosidases activity.
Keywords: Cellulosic biomass; Cellulase; β-glucosidase; Bacillus subtilis; Fermentation
Introduction
Large quantities of lignocellulosic wastes are generated through forestry, agricultural practices and industrial processes, particularly from agro-allied industries such as breweries, paper-pulp, textile and timber industries [1]. These lignocellulosic wastes are causes of environmental pollution otherwise lignocellulosic wastes are major natural resources of the world and major resource of renewable organic matter. As lignocellulosic biomass is an abundant and inexpensive renewable energy resource [2], attention has been drawn towards production of platform molecules for new bioproducts by microbial fermentation from glucose or other simple carbohydrates. The plant biomass regarded as wastes are biodegradable and in natural course they are degraded by microorganisms. They can be converted into valuable bioproducts such as biofuels, chemicals, cheap energy sources for fermentations, improved animal feeds and human nutrients [3]. Thus, profitable biomass conversion processes are highly dependent on the use of efficient enzymes for lignocellulose degradation. Microorganisms bring about most of the cellulose degradation occurring in nature. They meet this challenge with the aid of a multi-enzyme system [4]. Cellulases are inducible enzymes synthesized by a large diversity of microorganisms including both fungi and bacteria during their growth on cellulosic materials [5]. They are composed of independently folding, structurally and functionally discrete units called domains or modules, making cellulases [6]. Cellulase is a family of at least 3 groups of enzymes, endo-(1, 4)-β-Dglucanase, exo-(1,4)-β-D-glucanase and β-glucosidases. Their action is considered to be synergistic as all three classes of cellulase can yield much more sugar than the addition of all three separately. Among the cellulolytic enzyme complex, β-glucosidase plays a key role and is the most critical that catalyze the final step in cellulose and hemicellulose into glucose [7]. It is generally responsible for the regulation of the whole cellulolytic process as it not only produces glucose from cellobiose, but also reduces cellobiose inhibition, allowing endocellulase and exocellulase enzymes to function more efficiently [8]. Thus, β-glucosidases allow the cellulolytic enzymes to function more efficiently by producing glucose from cellobiose and reducing cellobiose inhibition [9].
There has been much research aimed at obtaining new microorganisms capable of producing cellulolytic enzymes with higher specific activities and greater efficiency [10]. Cellulolytic enzymes play important role in natural biodegradation processes in which plant lignocellulosic materials are efficiently degraded by cellulolytic fungi, bacteria, actinomycetes, and protozoa. The conversion of cellulose to glucose is regarded as the rate limiting step in the production of biofuels and various valuable bioproducts from lignocellulosic materials, due to high cost of cellulases and their low efficiencies. So, identification, isolation and characterization of cellulase producing bacteria will continue to be an important aspect of biofuel research, biodegradation and bioremediation. The present research work is focused on presumptive identification of most efficient cellulase producing bacteria from biogas production plant effluent and dairy effluent based on morphological and biochemical properties and subsequent molecular characterization of the selected cellulase producing strains. Furthermore, identification of potential cellulolytic bacteria producing efficient β-glucosidaseby determining β- glucosidase activity.
Materials and Methods
This study was mainly conducted at the Bio-resource Technology and Industrial Biotechnology Laboratory, Department of Biotechnology and Genetic Engineering, Jahangirnagar University, Savar, Dhaka-1342, Bangladesh and Invent Technologies Ltd, Banani, Dhaka- 1213, Bangladesh.
Isolation, screening and identification of cellulolytic bacterial isolates
Isolation of cellulolytic bacteria: For isolating cellulolytic bacterial isolates, samples were collected from biogas production plant effluent and dairy effluent of dairy farm near Jahangirnagar University campus area by the standard sample collection procedure in a clean sterile plastic container and stored at 4°C until the analysis was carried out. For isolating cellulolytic bacteria 1 g of samples were first mixed with 10 ml of distilled water in a test tube and heated for 15 minutes at 85°C in hot water bath. After ten-fold serial dilution with saline water (0.85% NaCl), 0.1 ml of each dilution was spread on carboxyl methyl cellulose (CMC) agar plates under laminar airflow. Then the plates were incubated at 37°C for 24-48 hour in the incubator. Morphologically different bacterial colonies were purified by repeated streaking. The purified isolated single cells were preserved and aliquoted as stock ampoules in 40% glycerol solution and kept at -80°C for further identification and screening for cellulase production.
Primary screening of cellulolytic bacteria: For screening cellulolytic bacteria, purified bacterial isolates were inoculated into 5 ml of modified No.3 medium (3% Peptone, 1% CMC, 0.1% KH2PO4, 0.05%MgSO4, pH 6.8) and incubated at 37°C and 120 rpm for 24 hour in shaking water Bath. Then 5 μl of overnight grown culture of isolated bacterial cells were individually transferred in CMC agar plates and incubated at 37°C for 24-48 hour. After proper incubation, CMC agar plates were flooded with Grams iodine solution (2 g of KI and 1 g iodine in 300 ml distilled water) for 3-5 minutes and subsequently washed with 70% alcohol solution. After washing, only the cell producing an inhibition zonesare distinct, clear, and prominent zones of clearance around the colonies indicating cellulase production were selected.
Secondary screening and production of cellulase enzyme: The potential isolates selected by primary screening were then evaluated for enzyme production. Those isolates showing maximum CMCase activity were then considered for further studies.
Crude cellulase production: Newly isolated isolates were screened for cellulase enzyme production in submerged fermentation process. A basal media containg (2% Tryptone, 1% CMC, 0.1% KH2PO4, 0.1% K2HPO4, 0.04% MgSO4, 0.005% NaCl, and 0.000125% FeSO4, pH 7.0) was used for production of cellulase. For seed culture, a fresh isolated colony was inoculated in 5 ml basal media and incubated at 37°C and 120 rpm for 24 hour. The seed culture (5%) was then inoculated in 50 ml production media in a 250 ml conical flask and incubated at 37°C and 120 rpm for 24 hour in shaking water bath.
Preparation of crude enzyme: The cell free supernatant obtained by centrifugation served as crude enzyme source was used for determining the cellulase activity or further investigations. After 24 hour of incubation in shaking water bath, the cultures were centrifuged at 10000 rpm for 10 min using centrifuge to remove the unwanted material and supernatant was used as a source of crude enzyme. The obtained crude enzyme solution was utilized for determination of enzyme activities and stored at 4°C for subsequent use.
Determination of CMCase activity: The carboxy methyl cellulase (CMCase) activity was assayed using a method described by Miller [11], with some modifications [12]. For this assay, 500 μL of crude enzyme was added to 500 μL of substrate (1% CMC prepared in 50 mM sodium citrate buffer pH 5) in a test tube and incubated at 60°C for 30 min in the incubator. Two controls were run with the test sample. In the negative control, 500 μL of substrate was added with 500 μL of citrate buffer and in positive control 500 μL of standard cellulase enzyme was added to 500 μL substrate. The reaction was terminated by adding 2.0 ml of dinitrosalicylic acid (DNS) reagent and subsequently placing the reaction tubes in a water bath at 90°C for 15 minutes. One ml of 40% Rochelle salt (sodium potassium tartrate) solution was then added to the test tubes and kept in ice for 2 minutes to stabilize the color. The absorbance was recorded at 575 nm wave length using UV/Vis Spectrophotometer (OPTIZEN POP, Mecasys, 5U4603-124019-00, Korea) against the negative control. One unit of CMCase activity was defined as the amount of enzyme that liberated 1 μmol of reducing sugar (glucose) in 1 min at 37°C and pH 7.0 [13]. Those isolates showing maximum CMCase activity were then selected for identification.
Identification of most efficient cellulase producing bacteria: Bacterial isolates producing significant clear zone on CMC agar as well as higher CMCase activity were identified based on cultural, morphological and biochemical characteristics. For this, a series of conventional biochemical tests were carried out for the identification of the genus of bacterial strains according to the Bergey’s manual of systemic bacteriology [14].
The molecular identification was also done by 16S rRNA gene sequencing. Two universal primer for 16s rRNA gene amplification were used for the purpose of PCR. 27F (AGAGTTTGATCMTGGCTCAG) was used as the forward primer while U1492R (GGTTACCTTGTTACGACTT) was used as the reverse primer. The 16s rRNA gene was sequenced using ABI 3700 Genetic Analyzer in the 1st Base Laboratory SdnBhd, Malaysia. The 16s rRNA gene sequences were aligned using BioEdit7.2 software. The sample sequences were analyzed using BLAST-NCBI.
Determination of β-glucosidase activity: Three bacterial isolates showing higher CMCase activity were used for assaying β-Glucosidase activity. β-Glucosidase activity was assayed using (pNPG) pnitrophenyl- β-D glucopyranoside as a substrate. A reaction mixture 500 μl containing 375 μl of culture supernatant (enzyme), 50 μl of pNPG (10 mM) as substrate and 75 μl sodium acetate buffer (2M, pH 5.0) was incubated at 50°C for 10 min in hot water bath. The reaction was terminated by addition of 1M Na2CO3. The developed yellow colour was measured at 410 nm using spectrophotometer. The amount of p-nitrophenol (PNP) released was quantified using the PNP standard curve. One unit of β-glucosidase activity was expressed as the amount of enzyme required to release 1 μmole of PNP per minute under the assay conditions.
Results and Discussion
Isolation and primary screening of cellulolytic bacteria
In this study, two samples were collected from two different sites and total 16 isolates were obtained as shown in Table 1. The resulting 16 isolates were then tested on CMC agar plate for cellulase activity.
| Sites | Sample no. | Total no. of isolates | Labeled as |
|---|---|---|---|
| Biogas plant effluent | 1 | 6 | B1-B6 |
| Dairy effluent | 2 | 10 | D1-D10 |
Table 1: Different sites selected for sample collection to identify cellulase producers.
The isolated bacterial colonies forming clear-zones after application of Gram’s iodine solution were selected as cellulase producers. Zone of CMC hydrolysis of the isolates are given in the Table 2. Among them 6 isolates gave the maximum ratio of clear zone diameter to colony diameter on the CMC agar plate as compared to plates cultured with the other strains. So, these 6 isolates are efficient cellulase producers. For confirming their cellulolytic activity all these 16 isolates were then analyzed for secondary screening by CMCase activity assay.
| S. no | Cell name | Colony diameter n (mm) | Zone diameter z (mm) | Ratio (z/n) (mm) |
|---|---|---|---|---|
| 1. | B1 | 4 | 10 | 2.5 |
| 2. | B2 | 5 | 9 | 1.8 |
| 3. | B3 | 6 | 10 | 1.6 |
| 4. | B4 | 6 | 15 | 1.83 |
| 5. | B5 | 5 | 13 | 2.6 |
| 6. | B6 | 7 | 15 | 2.14 |
| 7. | D1 | 4 | 9 | 2.25 |
| 8. | D2 | 7 | 15 | 2.10 |
| 9. | D3 | 4 | 8 | 2.00 |
| 10. | D4 | 6 | 14 | 2.33 |
| 11. | D5 | 5 | 9 | 1.80 |
| 12. | D1 | 6 | 10 | 1.60 |
| 13. | D7 | 4 | 8 | 2.00 |
| 14. | D8 | 8 | 11 | 1.37 |
| 15. | D9 | 4 | 5 | 1.25 |
| 16. | D10 | 5 | 6 | 1.20 |
Table 2: Zone of CMC hydrolysis of different isolates (n ≥ 3).
Secondary screening and production of cellulase enzyme
On the basis of primary screening the potential isolates were then evaluated for their enzyme productivity in submerge fermentation process. CMCase activity assay for all the 16 bacterial isolates were carried out for secondary screening.
Determination of CMCase activity
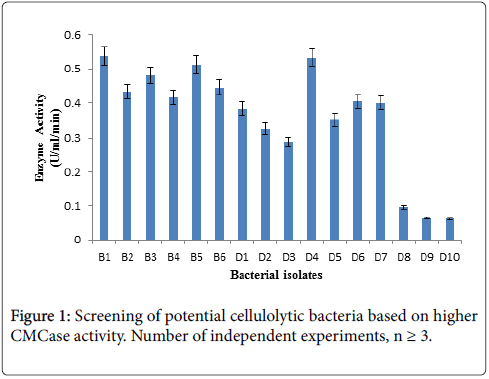
For this enzyme activity study, crude enzyme samples were assayed using 1% CMC as a substrate by DNS method. Three bacterial isolates giving higher absorbance at 575 nm in Figure 1 CMCase assay were selected as efficient cellulase producer.
Bacterial isolates (B1, B5, D4) producing significant clear zone on CMC agar as well as higher CMCase activity were selected and identified based on biochemical and molecular characteristics for further studies.
Identification of most efficient cellulase producing bacteria
Observation of biochemical characteristics: The biochemical identification of the selected isolates was performed according to the Bergey’s manual of systemic bacteriology [14] and all of the isolates were presumptively identified as Bacillus sp. The biochemical test results are presented in Table 3.
| Biochemical test | B1 | B5 | D4 |
|---|---|---|---|
| Gram staining | + | + | + |
| Shape | Rod | Rod | Rod |
| Methyl Red (MR) Test | -Ve | -Ve | -Ve |
| Voges-Proskaur (VP) Test | +Ve | +Ve | +Ve |
| Catalase Test | +Ve | +Ve | +Ve |
| Urease Test | +Ve | +Ve | +Ve |
| Oxidase Test | -Ve | -Ve | -Ve |
| Indole Test | -Ve | -Ve | -Ve |
| Citrate Test | -Ve | -Ve | -Ve |
| Gelatin Test | +Ve | +Ve | +Ve |
| Carbohydrate Fermentation Test | |||
| Glucose | +Ve | +Ve | +Ve |
| Fructose | +Ve | +Ve | +Ve |
| Sucrose | +Ve | +Ve | +Ve |
| Maltose | +Ve | -Ve | +Ve |
| Mannitol | +Ve | +Ve | +Ve |
| Finally identified bacteria | Bacillus sp. | Bacillus sp. | Bacillus sp. |
Table 3: Results of different morphological and biochemical tests on the isolates.
Observation of molecular characteristics:
DNA quantification and PCR: From B1, B5 and D4 bacterial isolates genomic DNA was extracted and quantified by Nanodrop Spectrophotometer and PCR was done by Takara PCR thermal cycler. The result of the extracted DNA quantification is presented in Table 4.
| Cell | B1 | B5 | D4 |
|---|---|---|---|
| Concentration of DNA (ng/µl) | 82.6 | 183.1 | 29.5 |
| A 260 | 1.652 | 3.661 | 0.59 |
| A 280 | 0.883 | 1.987 | 0.316 |
| A 260/280 | 1.87 | 1.84 | 1.87 |
| A 260/230 | 1.07 | 1.49 | 0.91 |
Table 4: DNA quantification result.
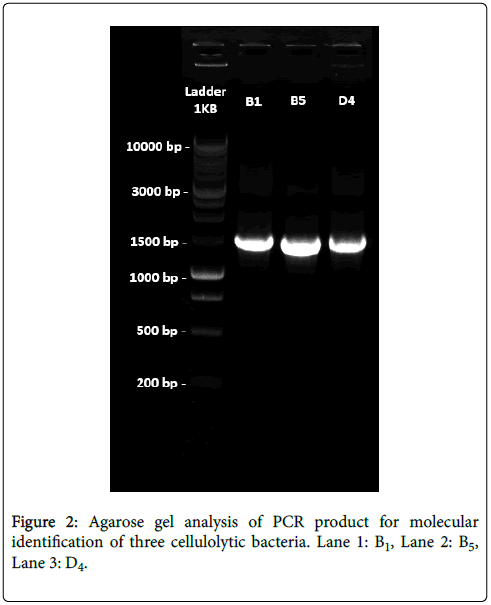
Agarose gel analysis for genomic DNA: After amplification of the extracted genomic DNA from the three cellulolytic bacterial strain agarose gel electrophoresis was performed using 1 Kb DNA ladder as a marker DNA for gel analysis. Three bands were found for our isolated bacterial DNA in the gel which were nearly 1.5 Kb (1500 bp) in comparing with 1 Kb DNA Ladder (Marker DNA).
16S rRNA gene sequences analysis: For all the three strains, the forward and the reverse sequence were analyzed. The nucleotide sequences coding for the 16S rRNA gene after BLAST query revealed that this gene is 100% homologous to Bacillus subtilis (B1), 99% homologous to Bacillus amyloliquefaciens (B5) and 100% homologous to Bacillus subtilis (D4) (Figure 2).
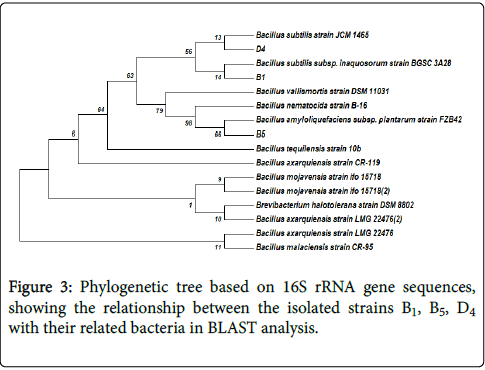
Phylogenetic analysis: A phylogenetic tree based on the blast search was constructed. Figure 3 shows the phylogenetic tree for 16S rDNA gene sequences.
Evolutionary analyses were conducted in MEGA7. The evolutionary history was inferred using the Neighbor-Joining method. The bootstrap consensus tree inferred from 1000 replicates was taken to represent the evolutionary history of the taxa analyzed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) was shown next to the branches. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. The analysis involved 18 nucleotide sequences Three cellulolytic bacteria isolated in this study were, Bacillus subtilis (B1), Bacillus amyloliquifaciens (B5) and Bacillus subtilis (D4), based on phylogenetic analysis of 16S rDNA sequence.
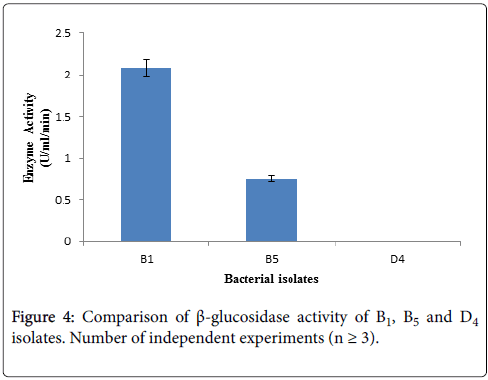
Determination of β-glucosidase activity: β-glucosidase activity assay was carried out with culture supernatant of three bacterial isolates having higher CMCase activity by using 10 mM pNPG as substrate. After terminating the reaction, absorbance was taken at 410 nm using spectrophotometer. Among the three selected cellulolytic bacterial strains, B1 showed higher absorbance thus it can produce β- glucosidase where D4 showed negative result thus it does not have β- glucosidase activity showing in Figure 4. Within these two strain B1 produce higher amount of β-glucosidase than B5 according to Figure 4.
The identified bacterial isolates B1, B5 and D4 all are Bacillus , due to their different environmental origin of sources they produce cellulytic enzymes depending on their own needs. Both B1 and D4 bacterial isolates are Bacillus subtilis , where B1 was isolated from biogas effluent plant and this source is enriched with municipal cellulolytic waste materials. So, these microbes produce higher amount of β-glucosidase due to some signal induction provided from their sourrounding environment to fulfill their requirement by utilizing cellulolytic wastes as their nutrients. On the other hand, D4 bacterial isolate was isolated form dairy effluent which is enriched with dairy waste materials. These microbes cannot produce β-glucosidase because they are not induced from their surroundings to produce this enzyme. Instead of being Bacillus subtilis , both B1 and D4 cannot produce β-glucosidase due to their different environmental origin of sources. As B1 bacterial isolate can produce significant amount of β-glucosidase, therefore further studies will be carried out with this B1.
Conclusion
Cellulase research has been concentrated mostly in fungi but there is increasing interest in cellulase production by bacteria due to their higher growth rate, thermo stable and alkali stable properties. The greatest potential importance is the ease with which bacteria can be genetically engineered. So, development of rapid and reliable methods for the screening of cellulases from microorganisms within inhospitable environments will allow a greater number of novel bacterial cellulases to be isolated with purpose for industrial use.
Moreover, most of the bioconversion processes used today does not allow complete saccharification of biomass. Hydrolysis of biomass can be enhanced by several approaches, one of which is by supplementation of cellulase complex with accessory enzymes like β- glucosidase. Our current knowledge of the production, purification, characterization, biochemistry, molecular biology of these enzymes and of their producer bacteria may help in this regard. Therefore, more advance research is required for a deeper understanding about these bacterial strains to improve their enzymatic production, purification and proper application of the enzyme from the isolate.
Acknowledgement
This research paper is partially supported by Ministry of Science and Technology, Bangladesh and INSPIRE program, British Council, Bangladesh.
References
- Abu EA, Onyenekwe PC, Ameh DA, Agbaji AS, Ado SA (2000) Cellulase (EC 3.2. 1.3) production from sorghum bran by Aspergillusniger SL1: An assessment of pretreatment methods. In: Proceedings of the international conference on biotechnology: commercialization and food security, Abuja, Nigeria pp: 153-157.
- Knauf M, Moniruzzaman M (2004) Lignocellulosic biomass processing: a perspective. International Sugar Journal 106: 147-150.
- Howard RL, Abotsi E, Van Rensburg EJ, Howard S (2003) Lignocellulose biotechnology: issues of bioconversion and enzyme production. African Journal of Biotechnology 2: 602-619.
- Aubert JP, Beguin P, Millet J (1987) Biochemistry and Genetics of cellulose Degradation, Fungal and Bacterial enzyme systems and their manipulation. Academic Press, New York, USA pp: 11-30.
- Koo YM (2001) Pilot-scale production of cellulase using Trichodermareesei Rut C-30 in fed-batch mode. Journal of Microbiology and Biotechnology 11: 229-233.
- Henrissat B, Teeri TT, Warren RAJ (1998) A scheme for designating enzymes that hydrolyse the polysaccharides in the cell walls of plants. Febs Letters 425: 352-354.
- Bhat MK, Bhat S (1997) Cellulose degrading enzymes and their potential industrial applications. Biotechnology advances 15: 583-620.
- Harhangi HR, Steenbakkers PJ, Akhmanova A, Jetten MS, van der Drift C, et al. (2002) A highly expressed family 1 beta-glucosidase with transglycosylation capacity from the anaerobic fungus Piromyces sp. E2. Biochimica et BiophysicaActa (BBA)-Gene Structure and Expression, 1574: 293-303.
- Liu D, Zhang R, Yang X, Zhang Z, Song S, et al. (2012) Characterization of a thermostable β-glucosidase from Aspergillus fumigatus Z5, and its functional expression in Pichiapastoris X33. Microbial cell factories 11: 1-25.
- Sukumaran RK, Singhania RR, Pandey A (2005) Microbial cellulases-production, applications and challenges. Journal of Scientific and Industrial Research 64: 832.
- Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical chemistry 31: 426-428.
- Wood TM, Bhat KM (1988) Methods for measuring cellulase activities. Methods in enzymology 160: 87-112.
- Zhang YHP, Evans BR, Mielenz JR, Hopkins RC, Adams MW (2007) High-yield hydrogen production from starch and water by a synthetic enzymatic pathway. PloS one 2: e456.
- Sneath HA, Peter MS, Nicolas H, Hold GL (1986) Bergey's Manual of Systemic Bacteriology. Williams & Wilkins, Philadelphia, USA.
Citation: Neesa L, Jahan N, Noman Khann AA, Rahman MS (2017) Cellulolytic Bacillus may or may not Produce β-Glucosidase due to their Environmental Origin – A Case Study. J Bioremediat Biodegrad 8: 412 DOI: 10.4172/2155-6199.1000412
Copyright: © 2017 Neesa L, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 4732
- [From(publication date): 0-2017 - Apr 04, 2025]
- Breakdown by view type
- HTML page views: 3891
- PDF downloads: 841