Canine Distemper Virus Causes Apoptosis in HEK-293 Cells by both Extrinsic and Intrinsic Pathways
Received: 28-Oct-2022 / Manuscript No. cmb-22-78583 / Editor assigned: 31-Oct-2022 / PreQC No. `cmb-22-78583(PQ) / Reviewed: 11-Nov-2022 / QC No. cmb- 22-78583 / Revised: 18-Nov-2022 / Manuscript No. cmb-22-78583(R) / Accepted Date: 25-Nov-2022 / Published Date: 25-Nov-2022 DOI: 10.4172/1165-158X.1000249 QI No. / cmb- 22-78583
Abstract
Background: Apoptosis is a form of natural or stress induced cell death that plays a pivotal role in many cellular processes. Virus induced apoptosis is of significant importance since many viruses/viral proteins have been reported to induce apoptosis in different cell types. The present study was carried out to identify genes and pathways to explain the mechanisms involved in Canine Distemper Virus (CDV) induced apoptosis.
Method: For this, HEK-293 cells were infected with CDV-SH, a Snyder Hill strain of canine distemper virus, at different time points. Viability and apoptotic studies were performed using MTT and DNA laddering assays, respectively. qPCR arrays were custom designed to study the expression profile of 43 apoptotic genes in HEK-293 cells after 6, 12, 24 and 48hrs of CDV infection.
Results: MTT results showed 100%, 84.78%, 79.21% & 76.95% cell viability after 6, 12, 24 and 48hrs after infection. DNA laddering showed a faint laddering pattern at 24hr and 48hr post CDV infection which indicated small amounts of DNA fragmentation. Expression studies revealed increased expression of nineteen genes and down regulation of three genes in all the groups. Ingenuity Pathway Analysis (IPA) showed activation of ‘Apoptosis’ pathway along with significant upstream and downstream regulators in CDV infected HEK-293 cells.
Conclusion: Our study demonstrates that apoptosis could be detected in HEK-293 cell lines, as revealed by DNA laddering 24hrs post CDV infection. qPCR & IPA analysis revealed upregulation of caspase-8, caspase-9 and caspase-3 which showed the involvement of both extrinsic and intrinsic pathways of apoptosis in HEK-293 cells following CDV infection.
Keywords
Apoptosis; CDV; HEK-293 Cell Line; qPCR Array; Infection
Introduction
Apoptosis is a regulated form of cell death which occurs during physiological conditions. It plays a critical role in the homeostasis of multicellular organisms, and constitutes a common pathway for cell replacement, tissue remodeling, damaged cell removal and elimination of cancer cells. Cells undergoing apoptosis present typical morphological characteristics, including membrane blebbing, chromatin condensation, cell shrinkage and apoptotic body formation. Apoptosis is triggered by sequential activation of caspases, a group of cysteine proteases, and proceeds primarily through two pathways. The extrinsic pathway involves activation of caspase-8 and is initiated by ligand interaction with death receptors, while the intrinsic pathway is activated by an imbalance between proapoptotic and antiapoptotic proteins from Bcl-2 family in mitochondria. Many viral proteins can influence the cellular pathways that control cell proliferation and apoptosis. Some viral proteins trigger apoptotic cell death, and this may be important in host defense and viral spread. Many viruses have been identified to induce apoptosis in different cell types such as Newcastle Disease Virus (NDV), Measles Virus (MV), Influenza Virus, Herpes Simplex Virus, Vaccinia Virus [1].
Canine Distemper Virus (CDV) is an enveloped virus with a single stranded RNA genome belonging to the family Paramyxoviridae. The genome encodes eight proteins, two of which (V and C) are nonstructural proteins and are alternatively translated from the RNA and six structural proteins (large protein, nucleoprotein, haemagglutinin protein, fusion protein and matrix protein). It causes generalized infection with prominent respiratory, gastrointestinal and nervous signs and symptoms such as fever, cough, coryza and conjunctivitis. It has been observed that CDV infection induces apoptosis of different cell types such as Vero and Hela, characterized by changes in cellular morphology and biochemical features, including DNA fragmentation, cytoplasm vacuolation, plasma membrane blebbing, and apoptotic body formation.
It has also been reported that CDV induces apoptosis in cerebellum and lymphoid tissue of the infected dogs, and also in Vero cells. Many authors have demonstrated that CDV causes apoptosis in different cell types by evaluating the expression of the caspases such as only or few apoptotic proteins and concluding that CDV induced apoptosis is caused by either extrinsic or intrinsic pathway. However, apart from the classical pathways of apoptosis, many other pathways including different apoptotic genes/proteins have also been recently reported such as MAVS-MKK7-JNK2 pathway, ER stress induced pathway, RIG-1/MAVS pathway. Moreover, in order to deduce a potential canonical pathway for apoptosis in cell types induced by viruses, a detailed study of majority of the apoptotic proteins need to be done. To our knowledge, this is the first report of apoptosis by canine distemper virus in HEK-293 cell line. The aim of this study was to elucidate the detailed mechanism, gene(s)/protein(s) and pathways involved in CDV induced apoptosis in the HEK- 293 cells [2].
Materials and Methods
Culture of Cell lines and Infection with Canine Distemper Virus (CDV)
HEK-293 cells present in the laboratory were grown in DMEM media (Himedia) supplemented with 10% FBS, 2X Antibiotic- Antimyotic solution (Himedia) kept at 37°C with 5% CO2. HEK- 293 cells at 60-70% confluence were infected with CDV (Snyder Hill strain) passage-2 (ATCC, USA). After 4-5 days post infection, the cells were visualized for any observable cytopathic effects and were harvested. The harvested culture was used for confirmation of CDV growth by Reverse Transcriptase-PCR using diagnostic primers designed in the lab against the L-gene of CDV.
Cell Viability Assessment
The cell viability/survival percentage assay was performed using MTT assay. HEK-293 cells were grown in 96-well cell culture plates in 5 replicates and were infected with CDV-SH at specific hourly intervals (6, 12, 24 & 48 hrs) along with non-infected controls and plain media as blank. After the specific time interval of CDV infection (i.e. 6, 12, 24 & 48 hrs), 20 μl of yellow MTT (5 mg/ml in PBS) was added to each well including control and blank, wrapped in aluminium foil and kept in incubator at 37°C for 4 hrs. After 4 hrs, media with MTT was removed from the cells; 200 μl of acidic isopropanol (containing 0.04 N HCl) was added to wells including controls and pipetted up and down to mix the purple formazon cyrstals. The plate was kept at 37°C for 10 min. The absorbance was measured at 540 nm wavelength using 630 nm as the reference wavelength in BioTek microplate reader.
DNA Fragmentation Assay (DNA laddering)
HEK-293 cells were grown in 6-well cell culture plates and upon reaching 60-70% confluence, the cells were infected with CDV-SH p-2 at different time intervals (6, 12, 24 & 48 hrs) keeping one well as noninfected control. After the specific time interval, the cells were harvested by pipetting at 1500 rpm for 10 mins in swinging bucket rotor at room temperature. The cells were washed twice with 1x PBS and tail lysis buffer (IM Tris-HCl, 500mM EDTA, 10% SDS, 5M NaCl) along with Proteinase K was added and incubated overnight at 56°C. Aqueous phase was separated using Phenol: Chloroform: Isoamyl Alcohol (PCI) with 3M sodium acetate by centrifugation at 10000 rpm for 10 min at 4°C. The aqueous phase was collected and 500ul of chloroform was added. The suspension was centrifuged at 10000 rpm for 10 min at 4°C. DNA was precipitated with 500 ul of Isopropanol by keeping overnight at -20°C. DNA pellets obtained were washed with 70% ethanol and airdried and dissolved in 50 ul of TE buffer (1M Tris-HCL, 0.5M EDTA). The DNA was run on a 1.5% agarose gel at 50V for 2-3 hours. DNA ladders were visualized under UV transilluminator [3].
Real time RT-PCR Array
For studying expression of apoptotic genes upregulated or downregulated in HEK-293 cells following CDV infection, a qPCR array (Qiagen) [48 x 2 format] was designed with the list of cellular genes (43 in number plus 5 controls = 48 in total) as in Table 1. Roughly HEK-293 cells were grown in 2 wells of five 6-well culture plates and upon reaching 70-80% confluence, the growth media was removed and a gentle washing was given using DMEM maintenance media. The cells were infected using CDV-SH (p-2, 100 ul) in duplicate wells in each plate according to the hourly interval (6, 12, 24 & 48 hrs) while keeping non-infected controls. RNA was isolated from the harvested cultures after appropriate time intervals using RNeasy Mini kit (Qiagen). RNA quality was checked and quantified using Nanodrop (Thermo Scientific). RNA corresponding to 1 μg (for each sample) was used to synthesis cDNA using RT2 First Strand Kit (Qiagen) by following the manufacturer’s protocol. Real time PCR was carried out according to manufacturer’s recommendations and following the MIQE guidelines in a total volume of 25 μl dispensed into each well of the 96-well array format (1350 μl of 2x RT2 S*YBR Green Mastermix, 102 μl of cDNA synthesis reaction and 1248 μl of RNase-free water). Reactions were run on a Step One Plus (Applied Biosystems, ABI) using standard thermal cycling parameters (95°C for 10min, 40 cycles of 15 s at 95°C, 1 min at 60°C to perform fluorescence data collection and continued melting curve analysis). The results were exported using the ABI software and the results in the form of Ct values were organised in Microsoft Excel for further analysis [4].
Results
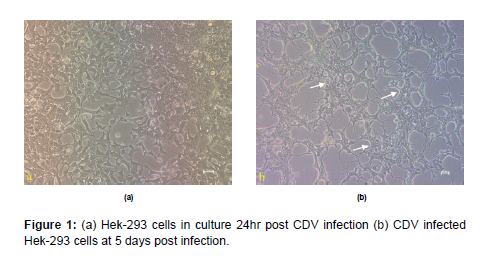
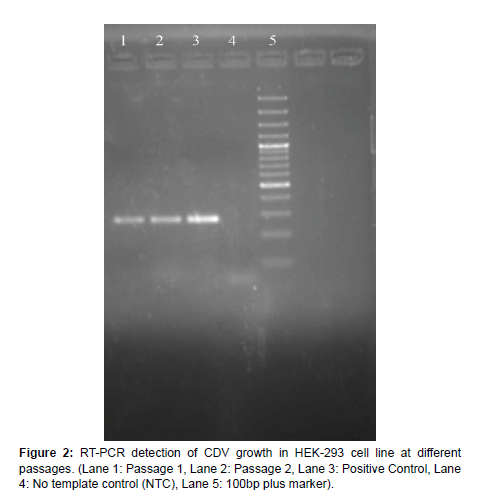
Culture of HEK-293 cells and infection with CDV HEK-293 cells available in the laboratory was grown in 25cc flasks using DMEM media supplemented with 5% FBS. At 60-70% confluence, the cells were infected with 100ul of CDV-SH, observed for 4-5 days and were harvested at 7 days’ post infection. Morphological changes in cells such as aggregation and degeneration could be observed without any characteristic CPE (Figure 1). Diagnostic RT-PCR could detect CDV growth in all passages which as observed by an amplicon size of 267 bp in agarose gel electrophoresis (Figure 2).
MTT Assay
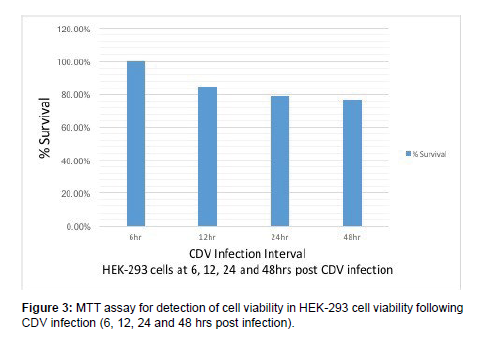
MTT assay was carried out in HEK-293 cells at 6, 12, 24, 48 hrs post CDV-SH infection. MTT assay showed 100%, 84.78%, 79.21% & 76.95% of cell viability in HEK-293 cells at 6, 12, 24 and 48 hrs post CDV infection respectively (Figure 3). Zhau reported decline of ovarian cancer cells survival following infection with live-attenuated measles vaccine virus in both time and dose dependent manner. The Measles virus vaccine resulted in 2.7%, 4.3% and 17.6% cell death at 12, 24 and 48 hrs post infection respectively in SKOV-3 cells by MTT assay.
DNA Laddering Assay
DNA isolated from CDV infected HEK-293 cells at 6, 12, 24 and 48 hrs post infection was electrophoresed in agarose gel at low voltage and a laddering pattern of DNA was observed. DNA laddering in HEK- 293 cells showed a faint laddering pattern at 24 & 48 hrs post CDV infection indicated small amounts of DNA fragmentation [5].
Gene expression studies following CDV infection in HEK- 293 cells using qPCR array
A panel of 48 genes associated with different apoptotic pathways listed in Table 1, were selected to study their expression profile following CDV infection in HCT-15 cells. Briefly, we assayed the expression of genes from the extrinsic pathway (5), genes related to the intrinsic pathway (10), genes from the MAVS-MKK7-JNK2 pathway (4), genes from RIG-1/MAVS pathway (4), execution pathway genes (3), genes from ER stress induced apoptosis pathway genes (4) and genes from PTEN induced pathway (2). The list of selected genes was sent to Qiagen for custom designing of the qPCR arrays. For apoptotic gene expression studies, HEK- 293 cells infected with CDV-SH were harvested at different time intervals (6, 12, 24 & 48 hrs). RNA was extracted and cDNA was prepared. A diagnostic RT-PCR was first carried out to detect CDV growth in the harvested samples which showed a 267 bp amplicon in agarose gel electrophoresis confirming the virus growth. Then the cDNAs synthesized from different samples were used to perform the qPCR array to study the apoptotic gene expression profile. The qPCR was performed in duplicates for each target gene and the mean Ct values obtained were recorded for analysis of the test samples (infection intervals) against the control sample (noninfected cells). The hourly infection intervals (6, 12, 24 & 48 hrs) were named into groups (group 1, 2, 3 & 4), respectively. The qPCR data was checked for quality control to check the PCR array reproducibility which shows that if the average PPC (PCR positive control) Ct is 20 ± 2 and no two arrays have average PPC Ct that are > 2 away from one another then the sample and group pass. Data normalization was done by manually selecting the housekeeping gene, and selecting the use of geometric or arithmetic mean by considering only genes with small changes in expression across different sample groups (differences in CT values less than 1). Fold change was calculated for relative quantification by comparing non- infected controls with infected samples of different time intervals. Fold-change values greater than one indicates a positive or an up-regulation and the fold-regulation is equal to the fold-change. Fold-change values less than one indicate a negative or down-regulation, and the fold- regulation is the negative inverse of the fold-change [6].
| 96-Well Custom PCR Array Template | ||
|---|---|---|
| # | Gene Symbol | Gene RefSeq # |
| 1 | Fas | NM_000043 |
| 2 | FasLG | NM_000639 |
| 3 | FADD | NM_003824 |
| 4 | Bid | NM_001196 |
| 5 | Bax | NM_004324 |
| 6 | Bak1 | NM_001188 |
| 7 | Bad | NM_004322 |
| 8 | Bcl-2 | NM_000633 |
| 9 | Cytochrome C (CYC1) | NM_018947 |
| 10 | APAF1 | NM_001160 |
| 11 | PARP1 | NM_001618 |
| 12 | MAVS | NM_020746 |
| 13 | Caspase-8 (CASP8) | NM_001228 |
| 14 | Caspase-9 (CASP9) | NM_001229 |
| 15 | Caspase-3 (CASP3) | NM_004346 |
| 16 | JNK1 (MAPK8) | NM_002750 |
| 17 | JNK2 (MAPK9) | NM_002752 |
| 18 | MKK7 (MAP2K7) | NM_145185 |
| 19 | PI3k (PIK3CA) | NM_006218 |
| 20 | mTOR | NM_004958 |
| 21 | IFN-a (IFNa1) | NM_024013 |
| 22 | IFN-b (IFNb1) | NM_002176 |
| 23 | RIG-1 (RARRES3) | NM_004585 |
| 24 | IRF-3 | NM_001571 |
| 25 | PTEN | NM_000314 |
| 26 | AKT1 | NM_005163 |
| 27 | TRAIL (TNFSF10) | NM_003810 |
| 28 | Smac (DIABLO) | NM_019887 |
| 29 | TNFa | NM_000594 |
| 30 | VDAC1 | NM_003374 |
| 31 | TRAF2 | NM_021138 |
| 32 | Noxa (PMAIP1) | NM_021127 |
| 33 | Puma (BBC3) | NM_014417 |
| 34 | Calnexin (CANX) | NM_001746 |
| 35 | Calreticulin (CALR) | NM_004343 |
| 36 | CHOP (DDIT3) | NM_004083 |
| 37 | Calpain (CAPNS2) | NM_032330 |
| 38 | ATF6 | NM_007348 |
The p values were calculated based on a student’s t-test of the replicate 2 ^ (-Delta CT) values for each gene in the control group and treatment groups. The fold change (cut off set to 2) was calculated for the upregulated and downregulated genes against the control (noninfected sample). The gene expression data was used to generate scatter plots of each group as compared to the control group (non-infected group). The scatter plot compares the normalized expression of every gene on the array between the two selected groups by plotting them against one another to quickly visualize large gene expression changes. The central line indicates unchanged gene expression. The dotted lines indicate the selected fold regulation threshold. Data points beyond the dotted lines in the upper left and lower right sections meet the selected fold regulation threshold [7].
The expression data was used to generate a heat map with cluster analysis of CDV infected HEK-293 cells. The clustergram in shows the qPCR array gene expression profile of CDV infected HEK-293 cells which shows group clustering analysis of differentially expressed genes identified in control versus CDV infected HEK-293 cells at specific time intervals (6, 12, 24 & 48 hrs). Table 2 shows the list of overexpressed genes in different groups and Table 3 shows the list of under expressed genes in the different groups.
S.No. |
GROUP 1 (6hr) | GROUP 2 (12hr) | GROUP 3 (24 hr) | GROUP 4 (48 hr) |
|---|---|---|---|---|
| 1 | TRAIL (TNFSF10) | TNF a | IFN-β (IFNb1) | TRAIL (TNFSF10) |
| 2 | TNF a | TRAIL (TNFSF10) | TRAIL (TNFSF10) | CHOP |
| 3 | TNF a | IFN-a(IFN-a1) | ||
| 4 | IRF-3 | RIG-1 (RARRES3) | ||
| 5 | Foxo3a | TNF a | ||
| 6 | Bak1 | Noxa (PMAIP1) | ||
| 7 | Puma | Caspase-9 | ||
| 8 | p53 | Caspase-3 | ||
| 9 | Cytochrome C (CYC1) |
Puma | ||
| 10 | Fas | |||
| 11 | Calreticulin (CALR) |
|||
| 12 | PERK (EIF2AK3) | |||
| 13 | IRE1 (ERN1) | |||
| 14 | JNK1 (MAPK8) | |||
| 15 | IRF-3 | |||
| 16 | IFN-β (IFNb1) | |||
| 17 | Calnexin (CANX) | |||
| 18 | FasLG | |||
| 19 | Bad | |||
| S.No. | GROUP 1 (6 hrs) |
GROUP 2 (12 hrs) | GROUP 3 (24 hrs) | GROUP 4 (48 hrs) |
|---|---|---|---|---|
| 1 | - | Calreticulin (CALR) | - | - |
| 2 | - | Caspase-8 | - | - |
| 3 | - | Smac | - | - |
Expression profile
The expression analysis of CDV infected HEK-293 cells at 6, 12, 24 and 48 hr post infection as compared with the non-infected controls, showed different genes that were overexpressed or under-expressed. In group 1 (6 hr PI), 2 genes (TRAIL & TNF α) were overexpressed and no genes were under-expressed. In group 2 (12 hr PI, 2 genes (TNFα & TRAIL) were overexpressed and 3 genes (Calreticulin, Caspase-8 & Smac) were under-expressed. In group 3 (24 hr PI), 9 genes (IFN- β, TRAIL, TNF α, IRF-3, Foxo3a, Bak1, Puma, p53 & cytochrome C) were overexpressed and no genes were under-expressed. In group 4 (48 hr PI) vs. control group, 19 genes (TRAIL, CHOP, IFN-α, RIG-1, TNF α, Noxa, Caspase-9, Caspase-3, Puma, Fas, Calreticulin, PERK, IRE1, JNK1, IRF-3, IFN-β, Calnexin, FasLG & Bad) were overexpressed and no genes were under-expressed [8, 9].
Analysis of CDV infected HEK-293 cells using IPA
To investigate the possible biological interactions of the differently expressed genes upon CDV infection of HEK-293 cells, all the datasets as described in with expression fold change values obtained from the qPCR array analysis studies were imported into the IPA tool. The list of differentially expressed genes analyzed by IPA revealed 12 significant networks and among them the top three networks identified by IPA were Death receptor signaling; Apoptotic signaling and Induction of apoptosis by HIV-1 comprising of 29 focused molecules and significance score of 27. Further, the IPA analysis also shows groups of differently expressed genes into biological mechanisms that are related to Cell Death and Survival 2.45E-04, Cellular Function and Maintenance 2.45 E-04, Cell Morphology 1.17 E-04 and Cellular Compromise 1.75 E-04. IPA also revealed in CDV infected HCT-15 cells the important upstream regulators are TP 53, Cisplatin & POU5F1. Since there were 4 groups (infection intervals) the data set was analyzed for each individual group compared to the control group in IPA. In group 1 (6 hr PI) fold change, the most important canonical pathway that was found was of the apoptotic signaling (p-value -6.27E-27) with a z-score of 3 and a ratio of 0.232 consisting of sixteen upregulated molecules. A graphical representation of the pathway with the molecules involved in apoptotic signaling was generated by IPA where (TNF/FasL, TNFR/Fas, Caspase 8, Bid, tBid, p53, JNK1, Bak, Bcl-2, Bad, Diablo, Cytochrome C, Caspase 9, Caspase 3, CAD, PARP) no. of genes were involved in sequential activation of each downstream molecule leading to caspase activation. In group 2 (12 hr PI) fold change, the apoptotic signaling was found to be significant (p-value -3.74E-15) with a z- score of 1.667 and a ratio of 0.13 consisting of nine upregulated molecules. IPA generated a graphical representation of the pathway with the molecules involved in apoptotic signaling in the group 2 (12 hr pi) cells where (TNF/FasL, TNFR/Fas, p53, Bak, Bcl-2, Caspase 9, Caspase 3, PARP) no. of genes were found to be upregulated and directly participated in the process of apoptosis. In group 3 (24hr PI) fold change, the apoptotic signaling was found to be significant (p-value- 1.69E-25) with a z-score of 3.5 and a ratio of 0.232 consisting of sixteen upregulated molecules. IPA showed a graphical representation of the molecules involved in apoptotic signaling pathway where (TNF/FasL, TNFR/Fas, Caspase 8, MKK4/7, JNK1, Bak, Bcl-2, Bad, Bax, Cytochrome C, Caspase 9, Caspase 3, CAD) no. of genes were found upregulated and interacted in a sequential manner leading to activation of downstream caspases. In group 4 (48hr PI) fold change, the significant canonical pathway was the apoptotic signaling (p-value -7.18E-30) with a z-score of 3.3 and a ratio of 0.261 consisting of eighteen upregulated molecules. A graphical representation of the pathway with the molecules involved in apoptotic signaling was generated by IPA where (TNF/FasL, TNFR/Fas, Caspase 8, Bid, tBid, p53, JNK1, Bak, Bcl-2, Bax, Diablo, Cytochrome C, Caspase 9, Caspase 3, PARP) no. of genes were found to be upregulated and influenced the subsequent molecules for execution of apoptosis. Further, the upstream regulator analysis showed that TP53, POU5F1, Cisplatin, Foxo3a are the important upstream regulator in all the groups with the possible targets and prediction activation states as shown by evidence for effects. The analysis of possible genes also revealed many downstream effects which correlate with diseases or functions that are induced upon CDV infection of Hek-293 cells. The IPA showed the expression pattern of apoptotic genes similar to that of predicted activation states of up to nine ‘diseases and function annotation’ like “Apoptosis; Cell death; Apoptosis of tumor cell lines; Cell death of tumor cell lines; Necrosis; Cell death of carcinoma cell lines and Degradation of DNA” with a higher p-value and a z-score in the CDV infected HEK-293 cells. Further, in group 1 the downstream effects analysis showed that 23 out of 27 genes have the measurement direction consistent with the increase in "Apoptosis". In group 2, the downstream effects analysis showed that 13 out of 16 genes have the measurement direction consistent with the increase in "Apoptosis". In group 3, the downstream effects analysis showed that 26 out of 30 genes have the measurement direction consistent with the increase in "Apoptosis". In group 4, the downstream effects analysis showed that 27 out of 29 genes have the measurement direction consistent with the increase in "Apoptosis". As described by IPA, the most highly rated network as shown is the “Apoptosis of Embryonic cell lines” with different genes that are predicted to increase and regulate the process of apoptosis [10-12].
Discussion
Prediction of apoptotic pathway activated upon CDV infection of HEK-293 cells
According to the list of overexpressed genes, in group 1 & 2 the expression of TNFα & TRAIL was seen to be upregulated. TNFα is a pro-inflammatory cytokine that is involved in plays a role in immune modulation, inflammation, viral replication. TNFα binds to cellular receptors such as TNFR1 and TNFR2 leading to conformational changes and binding to downstream death domains and activating death signaling. TNF-Related Apoptosis Inducing Ligand (TRAIL) is a trans membrane protein that belongs to the TNF family and is involved in the induction of apoptosis. TRAIL binds to receptors DR4 and DR5 which leads to its trimerization and clustering of the receptors intracellular domain which further activates the Death Inducing Signaling Complex (DISC). The trimerization causes its binding to the adapter molecule Fas Associated Death Domain (FADD) which leads to activation of caspase-8. Caspase-8 activation then cleaves caspase-3 which further initiates the cleavage of death substrates leading to cell death. Newcastle Disease Virus (NDV) infection causes an upregulation of TNF-α & TRAIL which induces the extrinsic apoptosis. Respiratory Syncytial Virus (RSV) causes a strong upregulation of TRAIL-R1, TRAIL-R2 & TRAIL in HepG2 cells which resulted in a susceptibility to TRAIL induced apoptosis. In group 3, in addition to TNFα and TRAIL, IFN-β was upregulated which is a signaling protein and acts as antiviral agents released or stimulated upon viral infection. The type IFNs including IFN-α & IFN-β are directly upregulated upon virus infection for e.g. influenza virus. Next, in our study the expression of Interferon Regulatory Factor (IRF-3) & Foxo3a was seen to be upregulated. IRFs are transcriptional regulators of IFNs and IFN inducible genes and also play a pivotal role in the regulation of immune responses. IRFs are induced upon virus infection and lead to the induction of IFNs, which act to inhibit the virus replication inside the cell. Besides its antiviral role, IRF-3 has been seen to induce apoptosis by the RLR induced pathway involving intracellular viral protein sensors RIG-1 & MDA5. Foxo proteins are forkhead family of transcription factors and Foxo3a protein functions as a tumor suppressor and a trigger for apoptosis via the upregulation of cell death associated genes such as Puma and Bim or the downregulation of anti-apoptotic proteins such as FLIP. The expression of Bak1, Puma, p53 & Cytochrome C was also upregulated in group 3 (24 hrs). Bak1 is a pro-apoptotic protein, which is an apoptosis regulator and is localized to the Mitochondrial Outer Membrane (MOM) and functions to induce the opening of Voltage Dependent Anion Channel (VDAC), leading to the release of Cytochrome C from the mitochondria. Foli observed the upregulation of Bak1, Bik & Bad in HIV infected patients. Puma (p53 upregulated modulator of apoptosis) is a pro-apoptotic protein whose expression is regulated by the tumor suppressor p53 gene. Puma is believed to interact with anti-apoptotic proteins Bcl-2 or Bcl-xl thus, stopping their interaction with Bax and Bak which then signals apoptosis to the mitochondria. Cytochrome C is a hemeprotein that is found associated to the inner membrane of the mitochondria and is involved in the initiation of apoptosis, where upon release into the cytoplasm, it binds to APAF1, further initiating the process of apoptosis by activation of caspases. In group 4, upregulation of 19 genes were observed which included most of the genes upregulated in group 3 and additionally, 4 proteins associated with endoplasmic reticulum stress induced proteins namely Calnexin, Calreticulin, Perk & IRE1 showed overexpression. Calnexin and Calreticulin function as molecular chaperons and help in folding and assembly of glycoproteins that pass through the endoplasmic reticulum. Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK) is located in the endoplasmic reticulum and functions to phosphorylate EIF2 which inactivates it leading to a stop in the general translation machinery of the cell. Inositol-Requiring Enzyme 1 (IRE1) has both kinase and endoribonuclease activity and is involved in endoplasmic reticulum stress induced mainly the Unfolded Protein Response (UPR). Group 4 also showed overexpression of caspase-9 and caspase-3, both belong to the family of caspases, which are cysteine-aspartate proteases which are involved in apoptosis induction. Apoptosis causes the release of cytochrome c from mitochondria and activation of apaf-1 (forming the apoptosome) which then cleaves the pro-enzyme of caspase-9 into the active dimer form. Caspase-3 is an executioner caspase which is cleaved by initiator caspases 8 or 9, which activates it leading to chromatin condensation and DNA fragmentation [13].
As observed by the gene expression analysis by qPCR array and Ingenuity Pathway database, it was found that in the group 1 (6 hrs PI), TNF α overexpression was observed which acts as a ligand for binding to the death receptor TNFR1. Downstream of the death receptor activation, activation of caspase-8 was observed which is an initiator caspase involved in extrinsic pathway. Upon ligand binding to death receptors, an adapter molecule FADD is recruited which binds to inactive caspase-8 thus forming a complex called Death Inducing Signaling Complex (DISC). The activated caspase further cleaves downstream caspases or associates in the cleavage of proapoptotic protein Bid, thus forming a cross-link with the intrinsic pathway of apoptosis. It was observed that in CDV infected HEK-293 cells, caspase- 8 led to the cleavage of Bid into tBid which further moves to the mitochondria leading to activation of Bcl-2 proapoptotic proteins. Bid, a BH3 only pro-apoptotic protein was found to be upregulated with the activation of tBid. It is known that following the death receptor pathway Bid is cleaved by caspase-8, this cleavage results in the formation of a truncated protein tBid. It is further suggested that tBid is capable of activating Mitochondrial Outer Membrane Permeabilization (MOMP) as a result of the imbalance in the pro and anti-apoptotic proteins that enables it to occur. Next, Bak activation was proposed in the pathway, which is Bcl-2 pro-apoptotic protein which localizes to the mitochondria and functions to induce apoptosis. The Bcl-2 proteins are a crucial checkpoint in the apoptosis at the mitochondria. During apoptosis, the BH3s such as tBid activate Bak and Bax, thus inducing cytochrome C release and finally leading to caspase activation. Further in the pathway analysis, the expression of Bax and Diablo was upregulated downstream of tBid and Bak. Upon apoptotic stimuli, it is believed that Bax and Bak get activated upon upstream signals and get oligomerized at the Mitochondrial Outer Membrane (MOM) and causes its permeabilization, leading to initiation of downstream apoptotic cascades. Thus, Bax and Bak work at appropriate levels to facilitate the release of cytochrome C and Smac/ Diablo into the inter membrane space. Direct IAP Binding Protein with Low Isoelectric Point (Diablo) release from the mitochondria has been shown to repress the inhibitor of apoptosis proteins and further activates caspase-9. As the pathway progressed the activation of Bad, Bcl-2 associated death promoter, which induces apoptosis by inhibiting anti-apoptotic proteins Bcl-2 & Bcl-xl. The expression of Bax, Diablo and cytochrome C was seen to be upregulated which describes the permeabilization of mitochondria with the release of Diablo. Cytochrome C is a heme protein which is associated with the inner membrane of mitochondria, which is involved in the initiation of apoptosis by its release from the mitochondria upon mitochondrial membrane permeabilization or sensing due to increase in intracellular calcium efflux. With the over- expression of proapoptotic proteins Bax, Bad & Bak, and the release of Diablo and cytochrome C from the mitochondria, there was upregulation of Apoptotic Protease Activating Factor (APAF1) which serves as a key molecule in the intrinsic apoptosis, which is known to oligomerize upon Cytochrome C release from the mitochondria resulting in the formation of a complex called the apoptosome. In this study, Downstream to APAF1 activation, activation of caspase-9 and caspase-3 was observed in CDV infected HEK-293 cells. It is known that activated APAF1 binds to the procaspase 9 to form the apoptosome complex through monomer interactions, which leads to the dimerization and formation of catalytically active caspase 9. Active caspase 9 leads to the cleavage and activation of the executioner caspases 3, 6 & 7 resulting in chromatin condensation, DNA fragmentation, cell shrinkage and finally apoptosis. IPA revealed the upregulation of major apoptotic proteins (Bak1, Bid, FasLG, MAP2K7, MAPK8, p53, Bcl-2, Cytochrome C, TNF, Bax, Diablo, Bad, APAF1, Caspase-8, Caspase-3, Caspase-9, PARP1) from both extrinsic and intrinsic pathway of apoptosis along with important upstream regulator and downstream regulators. As in this study, upregulation of caspase 8, 9 and 3 could detected which play major roles in the classical xtrinsic and intrinsic apoptotic pathways. Therefore, it is hypothesized that CDV induced apoptosis in HEK-293 cells occurs by both intrinsic and extrinsic pathways of apoptosis [14, 15].
Ethics Declarations
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed Consent
All the authors agree to submit for publication.
Acknowledgement
None
References
- Siegel RL, Miller KD, Jemal A (2019) Cancer statistics, 2019. CA Cancer J Clin 69: 7-34.
- Severson RK, Schenk M, Gurney JG, Weiss LS, Demers RY (1996) Increasing incidence of adenocarcinomas and carcinoid tumors of the small intestine in adults. Cancer Epidemiol Biomarkers Prev 5: 81-84.
- Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen YJ, et al. (2019) Small Bowel Adenocarcinoma, Version 1.2020, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 17: 1109-1133.
- Bilimoria KY, Bentrem DJ, Wayne JD, Ko CY, Bennett CL, et al. (2009) Small bowel cancer in the United States: changes in epidemiology, treatment, and survival over the last 20 years. Ann Surg 249: 63-71.
- Hatzaras I, Palesty JA, Abir F, Sullivan P, Kozol RA, et al. (2007) Small-bowel tumors: epidemiologic and clinical characteristics of 1260 cases from the connecticut tumor registry. Arch Surg 142: 229-235.
- Lepage C, Bouvier AM, Manfredi S, Dancourt V, Faivre J (2006) Incidence and management of primary malignant small bowel cancers: a well-defined French population study. Am J Gastroenterol 101: 2826-2832.
- Canavan C, Abrams R, Mayberry J (2006) Meta-analysis: colorectal and small bowel cancer risk in patients with Crohn's disease. Aliment Pharmacol Ther, 23: 1097-1104.
- Guth CA, Sodroski J (2014) Contribution of PDZD8 to stabilization of the human immunodeficiency virus type 1 capsid. J Virol 88: 4612-4623.
- Henning MS, Morham SG, Goff SP, Naghavi MH (2010) PDZD8 is a novel Gag-interacting factor that promotes retroviral infection. J Virol 84: 8990-8995.
- Henning MS, Stiedl P, Barry DS, McMahon R, Morham SG, et al. (2011) PDZD8 is a novel moesin-interacting cytoskeletal regulatory protein that suppresses infection by herpes simplex virus type 1. Virology 415: 114-121.
- Rampertab SD, Forde KA, Green PH (2003) Small bowel neoplasia in coeliac disease. Gut 52: 1211-1214.
- Chow JS, Chen CC, Ahsan H, Neugut AI (1996) A population-based study of the incidence of malignant small bowel tumours: SEER, 1973-1990. Int J Epidemiol 25: 722-728.
- Locher C, Batumona B, Afchain P, Carrere N, Samalin E, et al. (2018) Small bowel adenocarcinoma: French intergroup clinical practice guidelines for diagnosis, treatments and follow-up (SNFGE, FFCD, GERCOR, UNICANCER, SFCD, SFED, SFRO). Dig Liver Dis 50: 15-19.
- Algaba A, Guerra I, Marin-Jimenez I, Quintanilla E, Lopez-Serrano P, et al. (2015) Incidence, management, and course of cancer in patients with inflammatory bowel disease. J Crohns Colitis 9: 326-333.
- Zhang S, Sodroski J (2015) Efficient human immunodeficiency virus (HIV-1) infection of cells lacking PDZD8. Virology 481: 73-78.
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Citation: Singh S, Deka D, Grewal BS, Jairath M, Verma R (2022) Canine Distemper Virus Causes Apoptosis in HEK-293 Cells by both Extrinsic and Intrinsic Pathways. Cell Mol Biol, 68: 249. DOI: 10.4172/1165-158X.1000249
Copyright: © 2022 Singh S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 1378
- [From(publication date): 0-2022 - Apr 04, 2025]
- Breakdown by view type
- HTML page views: 1014
- PDF downloads: 364