Research Article Open Access
Candida parapsilosis Sensu Stricto: A Common Colonizing in Oral Mucosa of Argentine Subjects
Rodriguez L1,2*, Rosa A3 and Jewtuchowicz V41Department of Diagnostic Clinic and Semiology, School of Dentistry, Cuenca University, Ecuador
2Researcher for Mycology Center, IMPaM, Buenos Aires University – CONICET, Argentina
3Department of Microbiology and Parasitology, School of Dentistry, Buenos Aires University, Argentina
4Department of Microbiology, Parasitology and Immunology, Buenos Aires University, School of Medicine, IMPaM UBA-CONICET, Argentina
- Corresponding Author:
- Rodriguez L
Senior Teacher for the Department of Diagnostic Clinic and Semiology, School of Dentistry, Cuenca University , Ecuador
Tel: 5932933554
E-mail: malourdes84@hotmail.com
Received Date: November 02, 2016; Accepted Date: December 06, 2016; Published Date: December 15, 2016
Citation: Rodriguez L, Rosa A, Jewtuchowicz V (2016) Candida parapsilosis Sensu Stricto: A Common Colonizing in Oral Mucosa of Argentine Subjects. J Emerg Infect Dis 1:118. doi: 10.4172/2472-4998.1000118
Copyright: © 2016 Rodriguez L, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Infectious Disease and Pathology
Abstract
Candida parapsilosis is a complex made up of three species (Candida parapsilosis sensu stricto, C. orthopsilosis and C. metapsilosis) which differ genetically. In Argentina and worldwide, no data is available on the distribution and behavior of the complex in oral cavity niches, and there is little information on its epidemiology.
Aim: To characterize Argentine isolates of the C. parapsilosis complex from different oral cavity sites and other ecological niches.
Methodology: Retrospective, cross-sectional, descriptive study on a collection of isolates which were previously identified by conventional methods as parapsilosis complex, in order to distinguish the species by using end-point PCR with specific primers derived from a single sequence in the ITS1-5.8SrRNA-ITS2 region.
Results: 95% of the isolates were identified as Cp. sensu stricto, which was recovered with higher probability from oral mucosa sites, under pathological conditions, and in presence of intraoral appliances. Seventy-four percent of the strains were recovered under conditions of immunocompetence, and 100% of the isolates had resistant phenotype to flucytosine.
Conclusions: C. parapsilosis sensu stricto is a common species in different ecological niches. It is more likely to be recovered under conditions of immunocompetence. Dysbiosis of the mouth favors the growth of Cp. sensu stricto, which under these conditions may become a source for cross-transmission of more or less virulent strains by direct person-to-person contact, and a potential source of candidemia or invasive infections through hematogenous dissemination of strains with increased pathogenicity.
Keywords
Candida parapsilosis complex; Candida parapsilosis sensu stricto; Oral dysbiosis; Immunological status
Introduction
Results on the distribution of the species in this complex are highly variable, although all the literature we reviewed reports Candida parapsilosis sensu stricto as having the highest prevalence worldwide, and Silva et al. [1] report it as the most frequent isolate in hematogenous infections. The distribution of Candida orthopsilosis and Candida metapsilosis varies widely according to geographic region, clinical service and anatomical site [2,3]. Indeed, Miranda et al. [4] (claim that the exact importance of C. orthopsilosis and C. metapsilosis as human pathogens is as yet uncertain.
Little is known regarding the prevalence of species of the Candida parapsilosis complex in the oral cavity. The few papers published on the subject report variable results on its distribution in this specific ecological niche, and state that C. parapsilosis sensu stricto is the most commonly recovered species in immunocompetent individuals both in Europe and in North America [1,5,6] while C. metapsilosis seems to prevail over C. parapsilosis sensu stricto in immunocompromised patients [7].
Candida parapsilosis is usually susceptible to antifungal agents, but there are reports of isolates with reduced susceptibility to azoles and echinocandins [4]. This has serious clinical implications, since these are the drugs used as first-line agents for the treatment of invasive micosis [1,8].
Despite the importance of C. parapsilosis as a hospital pathogenic fungus, there are few studies, particularly in Latin America, on global epidemiology or on response to antifungal agents of species in this complex, and still less is known regarding its distribution in oral cavity niches. The oral cavity may be a source of candidemias caused by this fungus in patients with risk factors. In Argentina, there is very little information on the distribution and response to antifungal agents of the species in the complex, hence, our aim is to study the prevalence, distribution in the mouth and other ecological niches, and response to antifungal agents of the 3 species in the parapsilosis complex, from a collection of clinical isolates obtained in previous research studies and stored at the Mycological Center at the Buenos Aires University (UBA) School of Medicine.
Materials and Methods
This was a retrospective, cross-sectional, descriptive study using 150 clinical isolates of the parapsilosis complex from oral cavity and other ecological niches, stored in the collection at the IMPAM Mycology Center, School of Medicine, Buenos Aires University (UBA), which were collected during previous research projects from immunocompetent outpatients and hospitalized immunocompromised patients in different clinical situations.
The sample consisted of 101 isolates which were successfully reconstituted, to be analyzed by end-point PCR with specific primers. The following reference strains from the ATCC collection were used: C. parapsilosis (ATCC 22019), C. orthopsilosis (ATCC 96139) and C. metapsilosis (ATCC 96143), on which the same procedures were performed as on the clinical isolates.
For clinical correlation, patient´s clinical records and the dental data of the oral isolates were available.
For the in vitro susceptibility tests, we used Vitek2 automated susceptibility testing cards AST-YS07 to evaluate the response of 50 clinical isolates to the following antifungal agents: fluconazole (FLC), voriconazole (VRC), caspofungin (CASPO), micafungin (MICA) and amphotericin B (AMB). To interpret the readings, we used the 2012 revision of species-specific clinical breakpoints (CBPs) and epidemiological cutoff value (ECV) (Pfaller and Diekema 2012) [9-12] (CLSI, M27-S4/2012). For quality controls for the study we used the following Candida strains: Candida kruse iATCC 6258, C. parapsilosis ATCC 22019 and C. albicans ATCC 9002.
The following variables were analyzed: a) species of the parapsilosis complex; b) oral ecological niche; c) oral clinical status; d) intraoral appliances; e) immunological status; and f) response to antifungal agents.
Reconstitution of clinical isolates
Isolates were initially identified based on the color developed in the chromogenic medium, micro-morphology in 1%-Tween 80 milk agar and carbohydrate assimilation profile using commercial systems API ID 32D and Vitek2 (BioMérieux, France) [7,9].
To reconstitute the isolates, each strain was seeded in the following culture media: 1) BHI (brain-heart infusion) for metabolic activation of strains, incubated at 28°C-37°C for 24-48 h [7]; 2) Differential chromogenic solid medium for Candida (Chromagar), to ascertain the purity of the isolate and discard any contaminated strains, incubated at 28°C for 24 hours [10]; 3) Sabouraud, to amplify colonies, incubated at 28°C-37°C for 24 hours [10]; 4) YPD broth (yeast extract, peptone and glucose) to obtain a more robust culture, for 24 h with shaking at 37°C [10].
Molecular characterization of clinical isolates by end-point PCR with specific primers
Yeast DNA was obtained by breaking down the cell wall with zymolyase to generate spheroplasts (Scherer and Stevens 1987) [11]. Spheroplasts were verified by optical microscopy, after which a column (QUIAM Blood DNA Kit) was used for purifying, following the Qiagen protocol. The DNA obtained was preserved at -20°C.
Molecular typing was done by end-point PCR with specific primers derived from unique sequences contained in the internal transcriptional spacer 1 (ITS 1)-5.8 rRNA-(ITS2) of the fungal ribosomal DNA, which enable species-specific sequences for C. parapsilosis sensu stricto, C. orthopsilosis and C. metapsilosis to be retrieved separately (Table 1) [13,14].
| Primer | Target gene | Direction | Species specificity | Sequence | Amplification size |
|---|---|---|---|---|---|
| CPAF | ITS 1 | Forward | C. parapsilosis | TTTGCTTTGGTAGGCCTTCTA | 379pb |
| CPAR | ITS 2 | Reverse | GAGGTCGAATTTGGAAGAAGT | ||
| CORF | ITS 1 | Forward | C. orthopsilosis | TTTGGTGGCCCACGGCCT | 367pb |
| CORR | ITS 2 | Reverse | TGAGGTCGAATTTGGAAGAATT | ||
| CMEF | ITS 1 | Forward | C. methapsilosis | TTTGGTGGGCCCACGGCT | 374pb |
| CMER | ITS 2 | Reverse | GAGGTCGAATTTGGAAGAATGT |
Table 1: Primers used for rapid identification at species level of the C. parapsilosis complex. Source: Asadzadeh M, et al. (2009) [10].
Validation of PCR results with Sanger sequencing
The results of amplification with specific primers were validated with Sanger sequencing, for which an end-point PCR was performed using the pan-fungal primers ITS 1(forward: TCCGTAGGTGAACCTGCGG) and ITS 4 (reverse: TCTTTTCCTCCGCTTATTGATATG) to amplify and then sequence the ITS1-ITS4 region of ribosomal RNA gene 28S, as described by White et al. [2].
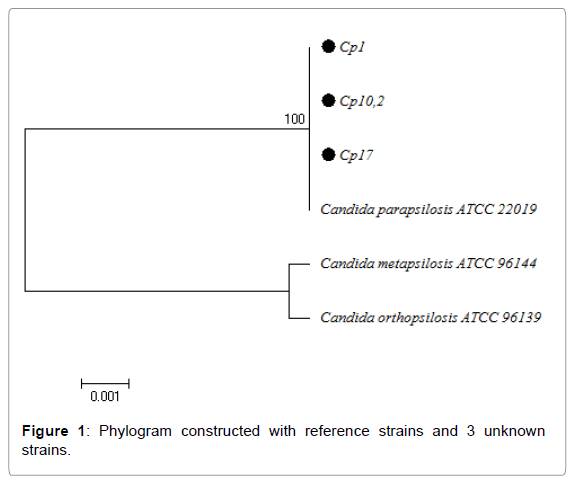
These primers amplified a 536 bp fragment from a fungal ribosomal DNA region (Figure 1). The PCR cycles were performed in a MiniCyclerTM, MJ Research INC thermal cycler, under the following protocol: one 5-minute cycle at 95º, followed by 30 3-stage cycles (20 seconds at 95°C//15 seconds at 55°C// 65 seconds at 72° C), and finally, one 5-minute cycle at 72°C [10].
The amplified fragments were purified using a commercial QIAquick PCR purification Kit (Qiagen), and sequenced using an ABI Prism 3730xl DNA analyzer (AppliedBiosystems, BsAs-Argentina) with the primers ITS1 and ITS4 [10].
Sequence identification and phylogenetic analysis
The sequences obtained were analyzed with the algorithm for sequence comparison BLAST (Basic Local AlignmentSearchTool)/ (http://www.ncbi.nlm.nih.gov/BLAST). To choose the best alignment between query and target sequence, we considered: a) % of identity; b) positive %; c) query coverage; d) and e-value.
For phylogenetic analysis we used the BIOEDIT software for editing sequence alignment and the MEGA 6 software for multiple alignment of sequences and phylogenetic analysis, for which the Neighbour joining algorithm was used. The tree was constructed with the reference ATCC sequences for C. parapsilosis, C. orthopsilosis, C. metapsilosis; in addition to the sequences selected at random from the total which were positive to PCR with specific primers.
Results
Of the total isolates upon which molecular analysis was performed, 96 (95%) were positive for the species C. parapsilosis sensu stricto (Table 2) according to the end-point PCR method with the pair of specific primers CPAR-CPAF, providing 379 bp amplicons (Figures 2-5). This band pattern is compatible to the one published by Asadzadeh et al. in the Journal of Medical Microbiology in 2009 [10]. The 5 remaining strains were negative for all three species of the parapsilosis complex, so they could only be identified by sequencing followed by bioinformatic analysis.
| Species | AF | RF | PF | CI95% |
|---|---|---|---|---|
| C. parapsilosis | 96 | 0.95 | 95% | 94.9-95% |
| C. orthopsilosis | 0 | 0 | 0 | 0 |
| C. metapsilosis | 0 | 0 | 0 | 0 |
| Others | 5 | 0.05 | 5% | 4.6-5.4% |
| Total | 101 | 1 | 100% |
Table 2: Distribution of frequencies for the species in the parapsilosis complex in the collection of clinical isolates.
DNA sequencing study in the ITS region
Of the 96 strains identified by PCR as C. parapsilosis, 3 strains were selected at random to validate by sequencing. The 5 strains that were negative for the 3 species of the C. parapsilosis complex were also sequenced to enable their identification.
The alignment determined 100% identical sequence for C. parapsilosis sensu stricto for all 3 strains (Cp1; Cp17; Cp10.2) selected at random, and which were positive for PCR for said species.
Phylogenetic analysis using MEGA 6 software confirmed the result obtained in BLAST, showing that the unknown strains and reference strain ATCC 22019 were 100% identical, with C. orthopsilosis and C. metapsilosis being more closely related to each other than they were to C. parapsilosis (Figure 1).
The BLAST analysis of the 5 strains that were negative according to PCR for the three strains in the complex determined the following identification:
Cp36A: 100% identical to C. albicans
Cp53.1: 100% identical to C. albicans
Cp78/2.2: 100% identical to C. pararugosa
Cp78PA: 100% identical to Kluyveromyces marxianus
Cp381: 100% identical to C. albicans
Prevalence and distribution of species in the parapsilosis complex according to ecological niche
Oral cavity: Thirty-eight isolates were successfully recovered from oral cavity at the following sites: tongue, cheek, palate, subgingival mucosa and peri-implant mucosa. Of the total C. parapsilosis strains isolated from mouth, 34 (89.5%) were positive for the species C. parapsilosis sensu stricto, while 4 were negative for the three species in the complex.
A higher frequency of isolation of C. parapsilosis was obtained in sites of oral mucosa with respect to subgingival niches. The difference was statistically significant and clinically relevant (Table 3).
| Type of oral niche | C. parapsilosis sensu strictoN(%) |
|---|---|
| Oral mucosa | 24 (70.6) |
| Gingival sulcus | 10 (29.4) |
| Total | 34 (100) |
PR=2.4 (CI95%: 1.7-3.1)
Table 3: Probability per ecological niche.
There is almost 4 times more probability of recovering C. parapsilosis of buccal cavity in pathological conditions than in health condition. The difference was statistically significant and clinically relevant (Table 4).
| Oral condition | C. parapsilosissensu stricto N(%) |
|---|---|
| With SF | 27 (79.4) |
| Without SF | 7 (20.6) |
| Total | 34 (100) |
Table 4: Probability per oral clinical condition.
The probability of recovering C. parapsilosis from the oral cavity with the presence of artifices is almost 3 times higher than in the absence of these. The difference was statistically significant and clinically relevant (Table 5).
| Intraoral appliances | C. parapsilosissensu stricto N(%) |
|---|---|
| With | 25 (73.5) |
| Without | 9 (26.5) |
| Total | 34(100) |
Fisher’s test: 0.000109359 (p: <0.05)
PR: 2.8 (IC95%: 2 – 3.6)
Table 5: Probability according to presence/absence of intraoral appliances.
All (100%) isolates from oral cavity were obtained from immunocompetent patients.
Other niches
Blood: 25 (25/101) isolates from patients with candidemias were analyzed, of which 24 (96%) were positive for the species C. parapsilosis sensu stricto. Only one isolate was negative for the three species in the complex, which was identified by sequencing as C. albicans.
Out of the total isolates from blood characterized as C. parapsilosis sensu stricto, 5 (20.8%) were from immunocompetent subjects and 19 (79.2%) from immunocompromised patients (Figure 4).
Skin and nails: 35 isolates were analyzed, of which 100% were identified as C. parapsilosis sensu stricto, with 31 (88.6%) coming from immunocompetent patients (Figures 4 and 5).
Urine: Two isolates were retrieved from urine, and confirmed as C. parapsilosis sensu stricto, one of which was from an immunocompetent and the other from an inmmunocompromised subject (Figure 4).
Rectal mucosa: Only one isolate was retrieved from rectal mucosa. It was identified as C. parapsilosis sensu stricto. It came from an immunocompromised patient (Figure 4).
C. parapsilosis was found to have a higher probability of being retrieved from immunocompetent than immunocompromised patients, with statistically significant difference (Table 6).
| Immunological status | C. parapsilosissensu stricto N(%) |
|---|---|
| Competent | 71 (74) |
| Compromised | 25 (26) |
| Total | 96 (100) |
PR= 2.84 (CI95%=2.34-3.34)
Table 6: Bivariate distribution of the parapsilosis complex according to host immunological status.
Antifungal susceptibility profile: Fifty of the 101 strains characterized as C. parapsilosis sensu stricto were selected to test for susceptibility to six antifungal agents which are regularly used in clinical practice: FLC, VRC, CASPO, MICA, AMB and FLUCY (Table 7). The selected strains came basically from the oral cavity and blood.
| Drug | S N(%) | IS N(%) | DDS N(%) | R N(%) | WT N(%) | noWT N(%) |
|---|---|---|---|---|---|---|
| FLC | 42 (84) | - | 5(10) | 3(6) | 42 (84) | 8 (16) |
| VRC | 49 (98) | 1(2) | - | - | 49 (98) | 1 (2) |
| CASPO | 48 (96) | - | - | 2(4) | 48 (96) | 2 (4) |
| MICA | 50 (100) | - | - | - | 50 (100) | 0 |
| AMB | ND | ND | ND | ND | 50 (100) | 0 |
| FLUCY | ND | ND | ND | ND | 0 | 50 (100) |
Table 7: Antifungal susceptibility profile of 50 strains of C. parapsilosis sensu stricto.
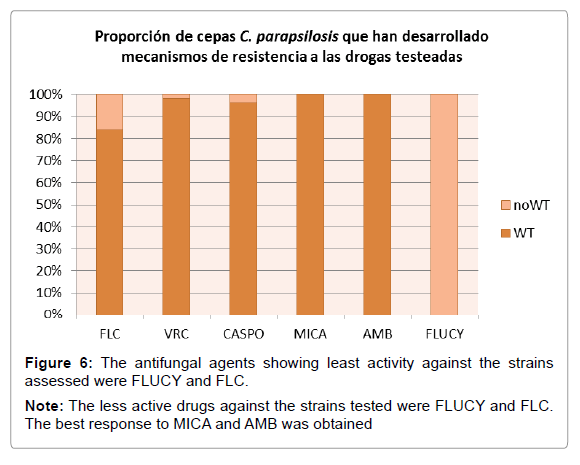
According to the species-specific cutoff point proposed by the CLSI Subcommittee in 2012, only 3 (6%) of the 50 strains showed a phenotype resistant to fluconazole, two of which were resistant to both FLC and CASPO. Five strains showed dose-dependent susceptibility to FLC (Table 7). It was not possible to classify the strains regarding amphotericin B because there is no validated clinical cutoff point. However, considering the epidemiological cutoff value (ECV) proposed for AMB and the species parapsilosis, 100% of the strains were wild-type phenotype (Figure 6). Since there is no validated CBP for flucytosine in this Candida species, the response of the 50 strains to it would be considered as undetermined (ND) (Table 7). However, considering the ECV, 100% of the strains were non wild-type to flucytosine, i.e. strains which have developed resistance mechanism to flucytosine (Figure 6).
Table 11 shows the minimum inhibitory concentration (MIC) needed to inhibit 50 and 90% of the strains, and the MIC range for each drug tested. The highest frequency of resistance and/or reduced susceptibility to FLC was obtained. For AMB and FLUCY, resistance frequency in the 50 strains is undetermined (ND) because there is no validated species-specific cutoff point. However, according to the epidemiological cutoff point (Pfaller and Diekema 2012) [12], all 50 strains were wild type to AMB and no wild type to FLUCY (Figure 6).
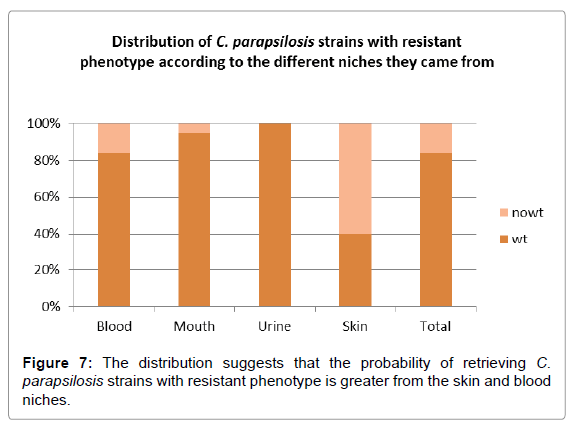
Analysis of the distribution of strains with resistant phenotype as determined by ECV, according to the niche from which they were isolated, showed that the largest percentage came from skin and blood (Figure 7).
Discussion
Many studies around the world report that within the complex, C. parapsilosis sensu stricto is the species most frequently recovered from clinical isolates in both pathological and commensal conditions [1-6,13,14]. However, there are few studies reporting prevalence and distribution of the species in this complex under conditions of oral health and disease. So far, C. parapsilosis sensu stricto is known to be the most prevalent species in oral cavity niches in conditions of immunocompetence, regardless of geographical region, as reported by studies from USA [5], Portugal [1], Turkey [6] and China [2]. One study from Brazil investigated the distribution of the species in this complex within the oral cavity of patients with chronic immunodeficiency due to HIV, in whom C. metapsilosis was the most frequently isolated species, followed by C. parapsilosis sensu stricto, although the difference was not statistically significant and the sample was very small [15].
In our study, only C. parapsilosis sensu stricto was isolated, with 100% prevalence in all samples recruited from oral cavity and other human ecological niches, predominating under conditions of immunocompetence. This result is similar to those reported by other authors such as Lotfali et al. [16] and Odds et al. [17-25], but in contrast to the results of most papers on the subject, which report recovery of C. orthopsilosis and C. metapsilosis in both hemocultures and various clinical samples (Tables 9 and 10).
| DRUG | MIC (ug/ml) |
CBPs | ECV | |||
|---|---|---|---|---|---|---|
| S | DDS/IS | R | WT | NoWT | ||
| FLC | 4 | x | X | |||
| VRC | x | X | ||||
| CASPO | x | X | ||||
Table 8: Response profile to FLC, VRC and CASPO for a strain from tongue which showed reduced susceptibility
| Ecological niche | C. parapsilosis | C. orthopsilosis | C. metapsilosis | Study country | Reference |
|---|---|---|---|---|---|
| Various clinical samples | 18 (90.0%) | 0 | 2 (10%) | Hungary | [18] |
| Hemocultures | 60 (95%) | 1(2%) | 2(3%) | Italy | [19] |
| Hemocultures | 67 (85.9%) | 5 (6.4%) | 6 (7.7%) | Spain | [20] |
| Various clinical samples | 111 (91.0%) | 10 (8.2%) | 1 (0.8%) | Spain | [21] |
| Invasive clinical samples | 1762 (92.1%) | 117 (6.1) | 34 (1.8%) | Global | [3] |
| Hemocultures | 29 (70.7%) | 10 (24.4%) | 2 (4.9%) | Malaysia | [22] |
| Various clinical samples | 109 (95.6%) | 5 (4.4%) | 0 | Kuwait | [10] |
| Variasmuestrasclínicas | 160 (94.6%) | 4 (2.4%) | 5 (2.9%) | Portugal | [1] |
| Hemocultures | 126 (88.1%) | 13 (9%) | 4 (2.8%) | Brazil | [13] |
| Hemocultures | 75 (95.0%) | 2 (2.5%) | 2 (2.5%) | Denmark | [23] |
| Invasive clinical samples | 112 (53.3%) | 30 (14.3%) | 56 (26.7%) | China | [24] |
| Various clinical samples | 81 (83.5%) | 7 (7.2%) | 9 (9.3%) | Taiwan | [25] |
| Various clinical samples | 41 (71.9%) | 0 | 16 (28.1%) | Eastern China | [2] |
| Various clinical samples | 96 (100%) | - | - | Argentina | This study |
Table 9: Distribution of species from the Candida parapsilosis complex, summarized from published papers
| Author | C. parapsilosis | C. orthopsilosis | C. metapsilosis | Study country | Reference |
|---|---|---|---|---|---|
| N (%) | N (%) | N(%) | |||
| Moris et al. | 7 (46.7) | 0 | 8 (53.3) | Brazil (2014) | [15] |
| Enger et al. | 9 (64.3) | 5 (35.7) | 0 | Global (2001) | [32] |
| Silva et al. | 65 (94.2) | - | 4 (5.8) | Portugal (2009) | [1] |
| This study | 34 (100) | 0 | 0 | Argentina | [33] |
Table 10: Distribution of C. parapsilosis complex species in oral cavity niches, summarized from published studies.
Our study found that the probability of recovering C. parapsilosis sensu stricto from oral cavity is higher under pathological conditions, in agreement with a Chilean study published in 2008 [26], which reports a lower prevalence of yeasts, as reflected by a lower count of colonyforming units (CFU/ml) of Candida species, among periodontally healthy subjects than among periodontally affected subjects, with statistically significant difference. We also found that C. parapsilosis is more often recovered in presence of prosthetic or orthodontic devices. This is consistent with Jewtuchowicz et al. [27] who found higher prevalence of Candida genus yeasts from subgingival niches in immunocompetent non-smokers with periodontal condition who wore prosthetic devices than in non-users, identifying both albicans and non-albicans species, and recovering C. parapsilosis in the latter group. Our study also found that colonization by C. parapsilosis sensu stricto is more common in oral mucosa than in gingival sulcus. This is in agreement with Urzúa et al. [26], who showed that it is more common to recover Candida parapsilosis from oral mucosa sites than from subgingival niches, in conditions of both health and disease; with C. albicans y C. dubliniensis being more frequently isolated from the subgingival niche [26]. It should be highlighted that there is no published paper specifically studying the parapsilosis complex in relation to the three variables analyzed. However, a review of PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) did show that to date, recovery of C. parapsilosis and other non-Candida albicans species such as C. tropicalis, C. dubliniensis and C. glabrata is greater in subjects with periodontal disease [26,28], poorly controlled diabetics [29]; female subjects who use oral contraceptives [30]; and male subjects who use androgenic steroids [31,32]. Based on our review of this database, we can say that ours is the first study in Argentina and the American continent to study the distribution and behavior of this complex in the oral cavity based on a collection of more than 20 isolates (Table 10).
Seventy-four percent of the strains came from immunocompetent subjects. This result is consistent with Constante et al. [33], who report that C. orthopsilosis is able to produce disease mainly in immunodepressed patients. C. parapsilosis may thus be the most pathogenic species in the complex, since it has shown that it is able to produce disease especially in conditions of immunocompetence.
Resistance or tolerance to antifungal agents in species in the parapsilosis complex is a growing problem in the context of usual and new antifungal agents. According to some studies, C. parapsilosis sensu stricto isolates are less susceptible than C. orthopsilosis and C. methapsilosis isolates to some antifungal agents used in the treatment of candidiasis, such as amphotericin (AMB), fluconazole (FLC), itraconazole (ITC) and caspofungin (CASPO). Among the azolederived agents analyzed in our study, FLC was the least active, with 8 C. parapsilosis sensu stricto strains showing resistance and/or reduced susceptibility to it. In agreement with this, Silva et al. [9] found an MIC for FLC equal to 16ug/mL for a C. parapsilosis sensu stricto isolate, and similar results have been reported by Gomez-Lopez et al. [34] In contrast, Ataides et al. [13] and Ruiz et al. [35] report resistance in a C. parapsilosis sensu stricto isolate to ITC but not to FLC. In this regard, Van Asbeck et al. [14] suggest that the differences in susceptibility to FLC may also reflect different affinities for azoles in the key enzyme that synthesizes ergosterol, 14-α-demethylase or for other enzymes in this pathway. Interestingly, MIC50 and MIC90 for both azoles (Table 11) found for the 50 study strains are comparatively higher than values reported for other regions such as Turkey [6], Spain [4], and Argentina [36]. This demonstrates geographic variability regarding the way in which species in this complex respond to antifungal drugs.
| DRUG | MIC50 (µg/ml) | MIC90 (µg/ml) | MIC RANGE (µg/ml) | RESISTANCE FREQUENCY |
|---|---|---|---|---|
| FLUCO | <=1 | 4 | <=1-64 | 16% |
| VORICO | <=0.12 | <=0.12 | <=0.12-<=0.25 | 2% |
| CASPO | 0.5 | 1 | 0.5->=4 | 4% |
| MICA | 0.5 | 1 | 0.5-1 | 0% |
| ANFO B | <=0.25 | 0.5 | 0.25-0.5 | ND |
| FLUCY | <=1 | <=1 | <=1 | ND |
Table 11: MIC50 and MIC90 values for each drug tested for the 50 study strains.
Two isolates were resistant to CASPO in our study. This is consistent with the worldwide trend, since many papers have reported that the MIC for CASPO in C. parapsilosis sensu stricto is higher than that for the other two species in the complex. Mutations in the FKS gene have been found to be associated with resistance to CASPO, as shown by the increase in MIC in mutant isolates compared to non-mutant or wildtypes [37,38]. In contrast to the results with CASPO; all 50 strains were sensitive to MICA, and the MIC50 and MIC90 for both echinocandins was comparatively lower than reported in other parts of the world such as Turkey [39,10], Spain [8]. But this response was similar to that reported by Lockhart [3] and Canton [40].
Based on ECV, all strains were susceptible to AMB, in agreement with other studies [16,18]. However, Ataides et al. [13] and Lockhart et al. [3], reported resistance of the species C. parapsilosis sensu stricto to AMB. The response of the parapsilosis complex species to AMB varies considerably among regions, which may be determined by intra-species genotype variability.
In our study, 100% of the isolates presented MIC values lower than or equal to 1ug/mL FLUCY. As with AMB, there is no consensus on the cutoff point for FLUCY for the species Candida parapsilosis. Nevertheless, considering the ECV classification proposed by Pfaller and Diekema in 2012 [12], we found that 100% of the strains were resistant to FLUCY, since according to ECV, an MIC value for FLUCY equal to or less than 0.5 μg/ml corresponds to a wild-type (WT) strain, while an MIC value greater than 0.5 μg/ml indicates a Non-WT strain which has developed resistance mechanisms. However, the British Society for Mycopathology [39] establishes as flucytosine-“sensitive” isolates those which have an MIC to flucytosine equal to or less than 1 μg/ml. Our study found an MIC value for flucytosine for the 50 strains which is higher than those reported by Silva et al. [1], Miranda et al. [4] and Cantón et al. [40] in Europe. But in India, Bhatt et al. [41] found a high resistance rate to flucytosine in C. parapsilosis isolates.
Due to the fact that we did not recover C. orthopsilosis or C. metapsilosis, we were unable to establish differences in the response profiles among the 3 species in this complex.
Conclusions
-- In Argentina, C. parapsilosis sensu stricto may be the most prevalent species in the complex in different human ecological niches, under conditions of both health and disease, being more likely to be retrieved in situations of immunocompetence.
-- C. parapsilosis sensu stricto is a common species in oral mucosa, predominating in pathological conditions and in presence of prosthetic devices.
-- In the Argentine population, C. orthopsilosis and C. metapsilosis are probably two uncommon species in different human ecological niches, under both pathological and commensal conditions.
-- Subgingival sites in the oral cavity may constitute an unfavorable microenvironment for colonization and development by C. parapsilosis.
-- The response of C. parapsilosis sensu stricto to antifungal agents seems to depend on the strain and the geographic region.
-- Mouth and skin may be reservoirs for C. parapsilsois strains with resistant phenotype and/or reduced susceptibility to the group of azoles, echinocandins and flucytosine.
Recommendations
Depending on the limitations of this study, we suggest:
- Repeating this study design but in a prospective model and with a larger sample size to minimize the random error inherent to the process.
- Validate the results of in vitro antifungal sensitivity with the reference method (CLSI M27-S4 / 2008)
- Evaluate sensitivity and specificity of the molecular technique employed in this study to discriminate at species level, by comparing it to the Gold standard in a large number of samples.
- Study the impact of the oral microenvironment in dysbiosis on the virulence of Candida parapsilosis sensu stricto.
References
- Silva A, Miranda I, Lisboa C (2009) Prevalence, Distribution, and Antifungal Susceptibility Profiles of Candida parapsilosis, C. orthopsilosis, and C. metapsilosis in a Tertiary Care Hospital. J Clin Microbiol 47: 2392-2397.
- Ge Y, Boekhout T, Zhan P (2012) Characterization of the Candida parapsilosis complex in East China: species distribution differs among cities. Med Mycol 50: 56-66.
- Lockhart S, Messer S, Pfaller M (2008) Geographic distribution and antifungal susceptibility of the newly described species Candida orthopsilosis and Candida metapsilosis in comparison to the closely related species Candida parapsilosis . J Clin Microbiol 46: 2659-2664.
- Miranda I, Eraso E, Hernandez J (2011) Prevalence and antifungal susceptibility patterns of new cryptic species inside the species complexes Candida parapsilosis and Candida glabrata among blood isolates from a Spanish tertiary hospital. J Antimicrob Chemother 66: 2315-2322.
- Ghannoum M, Jurevic R, Mukherjee P (2010) Characterization of the Oral Fungal Microbiome (Mycobiome) in Healthy Individuals. PLoS Pathogens 6: e1000713 .
- Tosun I, Akyuz Z, Guler N (2012) Distribution, virulence attributes and antifungal susceptibility patterns of Candida parapsilosis complex strains isolated from clinical samples. Med Mycol 51: 483-492.
- Mujica M, Finquelievich J, Jewtuchowicz V (2004) Prevalence of Candida albicans and Candida non-albicans in clinical samples during 1999-2001. Rev Argent Microbiol 36: 107-112.
- Pfaller M, Diekema D, Gibbs D (2008) Geographic and temporal trends in isolation and antifungal susceptibility of Candida parapsilosis: a global assessment from the ARTEMIS DISK Antifungal Surveillance Program, 2001 to 2005. J Clin Microbiol 46: 842-849.
- Trevino-Rangel R, Gonzalez-Gonzalez J, Garza-Gonzalez E (2012) Candida parapsilosis, una amenaza desafiante. Medicina Universitaria 14: 157-165.
- Asadzadeh M, Ahmad S, Al-Sweih N (2009) Rapid molecular differentiation and genotypic heterogeneity among Candida parapsilosis and Candida orthopsilosis strains isolated from clinical specimens in Kuwait. J Med Microbiol 58: 745-752.
- Scherer S, Stevens D (1987) Application of DNA typing methods to epidemiology and taxonomy of Candida species. J Clin Microbiol 25: 675-679.
- Pfaller M, Diekema D (2012) Progress in Antifungal Susceptibility Testing of Candida spp. by Use of Clinical and Laboratory Standards Institute Broth Microdilution Methods, 2010 to 2012. J Clin Microbiol 50: 2846-2856.
- Ataides F, Costa C, Hasimoto e Souza L (2015) Molecular identification and antifungal susceptibility profiles of Candida parapsilosis complex species isolated from culture collection of clinical samples. Rev Soc Bras Med Trop 48: 454-459.
- Van Asbeck E, Clemons K, Martínez M (2008) Significant differences in drug susceptibility among species in the Candida parapsilosis group. Diagnost Microbiol Infect Dis 62: 106-109.
- Moris D, Melhem M, Martins M (2012) Prevalence and antifungal susceptibility of Candida parapsilosis complex isolates collected from oral cavities of HIV-infected individuals. J Med Microbiol 61: 1758-1765.
- Lotfali E, Kordbacheh P, Mirhendi H (2016)Antifungal Susceptibility Analysis of Clinical Isolates of Candida parapsilosis in Iran. Iran J Public Health 45: 322-328.
- Odds F, Hanson M, Davidson M (2007) One year prospective survey of Candida bloodstream infections in Scotland. J Med Microbiol 56: 1066-1075.
- Kocsube S, Toth M, Vagvolgyi C (2007) Occurrence and genetic variability of Candida parapsilosis sensulato in Hungary. J Med Microbiol 56: 190-195.
- Barchiesi F, Orsetti E, Osimani P (2016) Factors related to outcome of bloodstream infections due to Candida parapsilosis complex. Barchiesi et al. BMC Infect Dis 16: 2-7.
- Gomez-Lopez A, Alastruey-Izquierdo A, Rodriguez D (2008) Prevalence and susceptibility profile of Candida metapsilosis and Candida orthopsilosis: results from population-based surveillance of candidemia in Spain. Antimicrob Agents Chemother 52: 1506-1509.
- de Toro M, Torres M, Maite R (2011) Characterization of Candida parapsilosis complex isolates. ClinMicrobiol Infect 17: 418-424.
- Tay S, Na S, Chong J (2009) Molecular differentiation and antifungal susceptibilities of Candida parapsilosis isolated from patients with bloodstream infections. J Med Microbiol 58: 185-191.
- Mirhendi H, Bruun B, Schonheyder H (2010) Molecular screening for Candida orthopsilosis and Candida metapsilosis among Danish Candida parapsilosis group blood culture isolates: proposal of a new RFLP profile for differentiation. J Med Microbiol 59: 414-420.
- Feng X, Wu Z, Ling B (2014) Identification and Differentiation of Candida parapsilosis Complex Species by Use of Exon-Primed Intron-Crossing PCR. J Clin Microbiol 52: 1758-1761.
- Chen Y, Lin Y, Chen K (2010) Molecular epidemiology and antifungal susceptibility of Candida parapsilosis sensustricto, Candida orthopsilosis, and Candida metapsilosis in Taiwan. Diagn Microbiol Infect Dis 68: 284-292.
- Urzua B, Hermosilla G, Gamonal J (2008) Yeast diversity in the oral microbiota of subjects with periodontitis: Candida albicans and Candida dubliniensis colonize the periodontal pockets. Med Mycol 46: 783-793.
- Jewtuchowicz V, Brusca M, Mujica M (2007) Subgingival distribution of yeast and their antifungal susceptibility in immunocompetent subjects with and without dental devices. Acta OdontolLatinoam 20: 17-22.
- Canabarro A, Valle C, Farias M (2013) Association of subgingival colonization of Candida albicans and other yeasts with severity of chronic periodontitis. J Periodontal Res 48: 428-432.
- Melton J, Redding S, Kirkpatrick W (2010) Recovery of Candida dubliniensis and other Candida species from the oral cavity of subjects with periodontitis who had well-controlled and poorly controlled type 2 diabetes: a pilot study. Spec Care dentista 30: 230-234.
- Brusca M, Rosa A, Albaina O (2010) The impact of oral contraceptives on women's periodontal health and the subgingival occurrence of aggressive periodontopathogens and Candida species. J Periodontol 81: 1010-1018.
- Brusca M, Verdugo F, Amighini C (2014) Anabolic steroids affect human periodontal health and microbiota. Clin Oral Investig 18: 1579-1586.
- Enger L, Joly S, Pujol C (2002) Cloning and characterization of a complex DNA fingerprinting probe for Candida parapsilosis. J Clin Microbiol 39: 658-669.
- Rodríguez L, Jewtuchowicz V (2016) Molecular characterization of Candida parapsilosis species complex in niches of the oral cavity in a cohort of patients from Argentina with different oral and dental clinical manifestations. J Dent Sci Ther1: 18-25.
- Constante C, Monteiro A, Alves A (2014) Different risk factors for candidemia occur for Candida species belonging to the C. parapsilosis complex. Med Mycol 52: 403-406.
- Gomez-Lopez A, Alastruey-Izquierdo A, Rodriguez D (2008) Prevalence and susceptibility profile of Candida metapsilosis and Candida orthopsilosis: results from population-based surveillance of candidemia in Spain. Antimicrob Agents Chemother 52: 1506-1509.
- Ruiz L, Khouri S, Hahn R (2013) Candidemia by species of the Candida parapsilosis complex in children's hospital: prevalence, biofilm production and antifungal susceptibility. Mycopathol 175: 231-239.
- Cordova S, Vivot W, Bosco-Borgeat M (2011) Sepecies distribution and susceptibility profile of yeast isolated from blood cultures: results of a multicenter active laboratory-based surveillance study in Argentina. Rev Argent Microbiol 43: 176-185.
- Pfaller M, Castanheira M, Diekema D (2010) Comparison of European Committee on Antimicrobial Susceptibility Testing (EUCAST) and Etest methods with the CLSI broth microdilution method for echinocandin susceptibility testing of Candida species. J Clin Microbiol 48:1592-1599.
- Cordoba S, Vivot W, Bosco-Borgeat M (2011) Species distribution and susceptibility profile of yeasts isolated from blood cultures: results of a multicenter active laboratory-based surveillance study in Argentina. Rev Argent Microbiol 43: 176-185.
- British Society for Mycopathology (1984) Laboratory methods for flucytosine (5-fluorocytosine). Report of a Working Group of the British Society for Mycopathology. J Antimicrob Chemother 14: 1-8.
- Canton E, Peman J, Quindos G (2011) Prospective Multicenter Study of the Epidemiology, Molecular Identification, and Antifungal Susceptibility of Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis Isolated from Patients with Candidemia. Antimicrob Agents Chemother 55: 5590-5596.
- Bhatt M, Sarangi G, Paty B (2015) Biofilm as a virulence marker in Candida species in Nosocomial blood stream infection and its correlation with antifungal resistance. Indian J Med Microbiol 33: 112-114.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 3601
- [From(publication date):
December-2016 - Apr 16, 2025] - Breakdown by view type
- HTML page views : 2718
- PDF downloads : 883