Mini Review Open Access
Bulked Segregant Analysis to Detect Main Effect of QTL Associated with Sheath Blight Resistance in BPT-5204/ARC10531 Rice (Oryza sativa L)
| Shailesh Yadav1*, Ghanta Anuradha1, Ravi Ranjan Kumar3, Vemireddy LR1, Ravuru Sudhakar2, Balram M1 and Siddiq EA1 | |
| 1Institute of Biotechnology, ANGRAU, Rajendranagar, Hyderabad-500030 India | |
| 2Seed Research and Technology Centre, Rajendranagar, Hyderabad-500030 India | |
| 3Department of Molecular Biology and Genetic Engineering, Bihar Agricultural University, Sabour-813210, India | |
| Corresponding Author : | Shailesh Yadav Institute of Biotechnology Acharya N G Ranga Agricultural University Rajendranagar, Hyderabad, India Tel: +91-7893473471 E-mail: shaileshagri9@gmail.com |
| Received: August 22, 2015; Accepted: September 28, 2015; Published: October 01, 2015 | |
| Citation: Yadav S, Anuradha G, Kumar RR, Vemireddy LR, Sudhakar R, et al. (2015) Bulked Segregant Analysis to Detect Main Effect of QTL Associated with Sheath Blight Resistance in BPT-5204/ARC10531 Rice (Oryza sativa L). J Rice Res 3:149. doi:10.4172/2375-4338.1000149 | |
| Copyright: © 2015 Yadav S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
| Related article at Pubmed, Scholar Google | |
| Keywords |
| ARC10531; Bulked segregant analysis (BSA); Rice; Sheath blight |
| Introduction |
| In the present scenario of increasing global human population, decreasing arable land, predicted increases of water scarcity, soil salinity, severe diseases, emerging resistance of pests and pathogens to pesticides and climate change pose significant challenges to modern rice research. The biotic stresses viz. , blast, stem rot, sheath blight, and bacterial blight diseases causes severe economic losses to rice productivity. Among them Sheath blight (ShB) is an important fungal disease caused by Rhizoctonia solani Kuhn causing up to 25% of yield loss and degrades rice quality. In hot and very high humid condition, yield loss can even reach as high as 50%. With the increasing application of nitrogenous fertilizers and the popularization of semi dwarf cultivars with more tillers, ShB is becoming the most serious disease in many rice-producing areas in the world [1-5]. The fungus R. solani Kuhn is soil borne pathogen which survives either as sclerotia or mycelia in plant debris. After the initial infection, the pathogen moves on the plant through surface hyphae and develops new infection structures over the entire plant, causing significant necrotic damage. The architecture of the canopy and the associated microclimate has strong effects on both the mobilization of primary inoculums and the further spread of the disease. Absolute resistance to R.solani is not available in any of the rice germplasm grown worldwide. However, it has been reported that resistance to R. solani is a typical quantitative trait controlled by polygenes in rice [6-12]. In rice because of availability of high resolution molecular maps, complete sequence information and extensive germplasm collections, mapping of quantitative trait loci (QTLs) for disease resistance such as sheath blight is feasible in crop improvement programme. In this context has reported for the first time the identification of rice QTL resistant to ShB using RFLP markers. To date, around 50 ShB resistance QTLs (ShBR QTLs) have been detected over all 12 rice chromosomes in cultivated varieties, deep-water varieties and wild species. Some of them were identified in multiple mapping populations and/or environments and not associated with either heading date (HD) or morphological traits, and they are believed to be stable ShB QTLs [9,10,12-14]. However QTL mapping is usually carried out by genotyping large number of progenies which is labor intensive, time consuming and cost-ineffective. Several strategies have been proposed to identify molecular markers near a gene/QTL of interest with reduced number of plants to be genotyped. The two main strategies are selective genotyping and bulk segregant analysis (BSA). Selective genotyping is relatively a low-cost approach to detect QTL with large effects by genotyping individuals from the two tails of the phenotypic distribution. Bulked Segregant Analysis (BSA) serves as an affordable strategy for mapping large effect QTLs by genotyping only the extreme phenotypes instead of the entire mapping population. BSA has been successfully used in rice for identifying markers linked to QTL associated with grain quality parameters blast resistance heat tolerance, drought tolerance gall midge resistance and sheath blight resistance. In the present study, bulked segregant analysis approach was used to identify large effect QTLs for sheath blight resistance and to observe the disease association with any of morphological traits [15-26]. |
| Materials and Methods |
| Plant materials |
| 40 rice germplasm lines including 8 wild, 4 land races, 26 cultivated and 2 advanced breeding lines were screened using typha bits method to identify the resistance source for sheath blight disease. A moderate level of resistance to this disease was identified in Tetep and ARC10531, a land race with the relative lesion height percentage of 21- 30%. Molecular mapping of QTLs using Tetep as a source for moderate resistance has already been carried out by several research groups. Hence ARC10531 was selected as male parent with highly susceptible elite variety BPT-5204.Crossing program and generation advancement to F2 was performed at the glass house facility, IBT, ANGRAU, Hyderabad during 2011 and 2012.The polymorphic SSR marker RM 205 and RM22565 were used to fix F1 progenies and true F1’s were forwarded to F2 generation [27,28] (Table 1). |
| DNA extraction and SSR analysis |
| Genomic DNA was isolated from frozen fresh leaf tissue of 210 F2:3 progenies along with both the parents (BPT-5204 and ARC10531) with the procedure described by [29]. The quality and quantity of DNA were estimated spectrophotometrically using a Nano Drop (ND-1000, Wilmington, USA).The final DNA concentration was adjusted to50 ng/ μl. Parental polymorphism survey involving 500 SSR markers spanning all twelve chromosomes was carried out. These SSR markers were selected based on uniform distribution across the 12 rice chromosomes. Out of the 500 SSR markers screened, seventy were found polymorphic among the parental lines (BPT- 5204 and ARC10531).The PCR reaction for SSR analysis was performed in volumes of \15 μl containing 50 ng genomic DNA, 0.2 μM each primers, 10 mM Tris-Hcl (pH 8.3), 50 mM KCl, 100 mM each of dATP, dGTP, dCTP and dTTP, 1.5 mM MgCl2 and 0.5 unit of Taq polymerase. The PCR amplification was performed on Eppendorf Mastercycler® Germany with a PCR profile of 94°C for 5 min. followed by 35 cycles of 1 min. at 94°C, 45 sec at 55°C and 1min at 72°C followed by final extension at 72°C for 10 min. The PCR products were separated on 3.5% metaphor-agarose gel and documented using a gel documentation system (BIORAD Gel Doctm XR, USA). |
| F2:3 phenotyping for sheath blight resistance and other attributes |
| The 210 individuals of F2:3 population of the cross (BPT-5204 × ARC10531) were inoculated with the local Rajendranagar isolate (AG1-IA) of Rhizoctonia solani which was obtained from Division of Plant Pathology, Directorate of Rice Research, Rajendranagar, Hyderabad-500030 in the year of 2011.The inoculums of virulent 00isolate were multiplied in Typha stem bits of 4-5 cm long soaked in typha medium (Peptone-10g, Sucrose- 20 g, K2HPO4-0.1 g, MgSO4-0.1 g dissolved in 1000 ml DDH2O pH6-6.5) [30]. Rice plants at maximum tillering stage were inoculated with R. solani by placing the typha pieces between tillers in the central region of rice hills 5-10 cm above the ground level. Rhizoctonia solani infected plants were kept in a humid chamber made of clear plastic for 2 weeks to allow disease development. Plants were grown at 28°C under 14-hrs day light in the humid chamber in the greenhouse. The humidity was maintained between 80 to 100% from the time of inoculation to disease evaluation. To evaluate sheath blight resistance or susceptibility of rice cultivars, the relative lesion height of inoculated plants were recorded 14 days after inoculation. The relative lesion height (RLH) was calculated by the following formula for scoring disease reaction: |
| Observations were recorded 14 days after inoculation and graded as per 0-9 Standard Evaluation System (SES) scale Following morphological attributes were recorded in segregating populations for a cross BPT-5204 × ARC-10531- Days to heading (days), plant height (cm), panicle length (cm), tiller no (number), effective tiller no (number), relative lesion height (percentage) [30]. |
| Marker-phenotype association analysis |
| Bulked segregant analysis (BSA) has been proposed as an efficient strategy for identifying DNA markers linked to the genes or genomic regions of interest [17]. Polymorphic markers may represent markers that are linked to a gene or QTL of interest [32]. DNA bulks of plants with extreme resistance and those with extreme susceptibility were prepared from F2:3 phenotyped progenies. This was done by pooling aliquots, containing equivalent amounts of total DNA approximately, 50 ng/μl from each of ten highly resistant and ten highly susceptible plants of the F2:3 based on phenotypic observations. 70 polymorphic SSR primer pairs were used for screening of parents and two bulk DNA samples. DNA of individual F2:3 plants that were included in bulks were also analyzed with co-segregating markers to confirm their linkage to the sheath blight disease resistance. The SSR markers found polymorphic among the parents and the bulks were used for F2:3 progeny analysis. DNA of 210 F2:3 progenies and parents were analyzed to study co- segregation of these markers. |
| Data Analysis |
| The clearly resolved amplicons of SSR were scored manually as homozygote for the allele for susceptible parent (B), homozygote for the allele for resistant parent (A) and heterozygote carrying the alleles from both parents (H) in the data sheet. Chi-square (χ2) test was performed to test the goodness of fit of the F2:3 population for the phenotyping and marker data by comparing an observed frequency distribution with an expected one. Marker-trait association was analyzed by simple linear regression method to know the association between the markers and the sheath blight score using software GenStatv14.1 Frequency distribution curve for sheath blight resistance of 210 F2:3 progenies were drawn separately using Microsoft Office 2010 Excel utility. Variability parameters were performed in F2:3 population viz. ,mean, range, skewness, kurtosis, standard deviation and simple correlation coefficients were worked out by software= GENSTAT v14.1[33]. |
| Results |
| Phenotyping of F2:3 progenies for sheath blight resistance and other morphological traits |
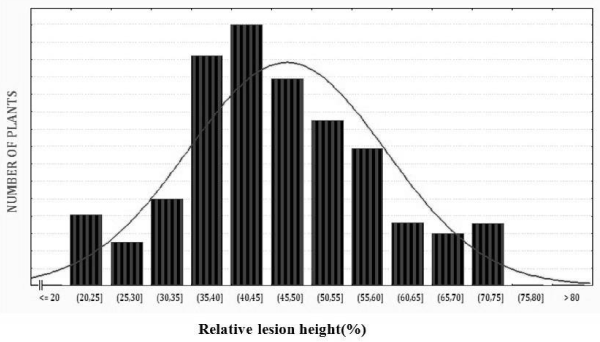
| The F2:3 progenies of the cross ARC10531 and BPT-5204 (210 progenies) were phenotyped for sheath blight during wet season 2012. The frequency distribution curve of F2:3 progenies for disease were continuous and near to normal distribution. The range of relative lesion height in percentage was 21-75%. In the F2:3 progenies, more individuals were distributed towards 40-50% of relative lesion height and population appears to be skewed more towards susceptible side. The mean value recorded for relative lesion height was 38.51%. The results revealed that variability for the morphological traits viz. , plant height, heading date , number of tillers, panicle length and disease score (RLH %) ranges from 70- 127, 69-33, 5-31, 5-25, 14-25 and 21- 75 respectively. The traits such as plant height, number of productive tillers and relative lesion height exhibit enough variability (Figure 1 and Table 2). |
| Rice microsatellite markers associated with sheath blight reaction using bulk segregant analysis: |
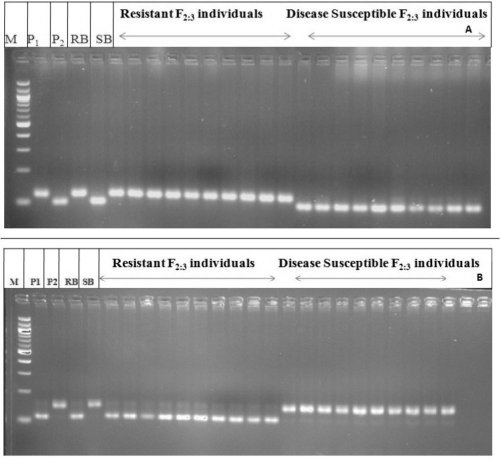
| Using the BSA method, two bulks having distinct and often contrasting phenotypes for the trait of interest are generated from a segregating population from a single cross. Seventy polymorphic markers were used for screening of parents ARC10531, BPT-5204, resistant bulk (RB), and susceptible bulk (SB) along with individuals of F2:3 populations used in respective bulks. Two markers RM 205 (chromosome#9) and RM 336 (chromosome#7) clearly distinguished susceptible bulks from resistant bulks. The F2:3 progenies were genotyped with these two primers (RM336 and RM205) to study their possible association with sheath blight resistance. Segregation pattern with marker RM336 recorded a resistant allele of donor in 43 plants, susceptible allele of recipient was amplified in 57 plants while 110 plants exhibited both the alleles (heterozygous). Similarly for marker RM205 observed 40 plants showing donor allele, 50 plants of recipient allele while remaining 120 plants depicted as heterozygotes. Genetic analysis with chi-square test indicated goodness of fit to the expected ratio of 1:2:1 for co-dominant marker indicating the association of RM336 and RM205 with sheath blight resistant gene in the present population. To determine the strength of association between the putative markers and the respective phenotypes, linear regression analysis was carried out using marker genotype as groups. The simple regression analysis between phenotypic data of sheath blight resistance and the genotypic data of SSR marker RM336 and RM 205 indicated that the marker was significantly linked with ShB resistance. The two tagged marker RM205 and RM 336 associated with ShB resistance was mapped too, using Mapmaker 3.0 (Cambridge, MA, USA) based on F2:3 genotyping data of all polymorphic markers identified on those two respective chromosomes. These results indicated the possible detection of two genetic loci for sheath blight resistance on chromosome 7 (based on map location of RM 336) and on chromosome 9 (based on map location of RM 205) (Tables 3 and 4) (Figures 2 and 3). |
| Discussion |
| Several groups have attempted to identify sources of ShB resistance by screening local accessions, cultivars, landraces, and/or advanced breeding lines. The genotypes which were most promising as sources of ShB resistance have been screened for ShB reaction in different rice growing regions [11]. Tetep is a well reported source of resistance to sheath blight and several QTLs have already been mapped by number of researchers [28,29]. The land race ARC 10531 identified in the present study was observed with equal levels of resistance as that of Tetep, used as an alternate source for ShB resistance in process of development of mapping population. The importance of land races in exploring valuable QTLs for sheath blight resistance is previously discussed by several researchers. The F2:3 progenies of the cross ARC10531 and BPT- 5204 were phenotyped for sheath blight during Kharif 2012 following standard method of screening in a hot and humid micro-chamber using the typha bits method reported by [31,34,35].The frequency distribution curve of F2:3 progenies for disease was continuous and near to normal distribution. Similar frequency distribution curve showing continuous variation involving the F2:3 progeny derived from Japonica cross was reported for ShB resistance. They also observed the curve depicting skewness towards susceptible reaction to disease [36]. Third and fourth degree statistics viz. , skewness and kurtosis of an F2:3 progenies suggests that as plant height is positively skewed (0.10) indicates that trait is associated with complementary interactions whereas heading dates (days) negatively skewed (-0.68) and associated with duplicate interaction. The remaining traits also exhibited positive skewness which is associated with complementary interactions. The skewed distribution of sheath blight trait in present study shows that it is under the control of non-additive gene action, especially epistasis and influenced by environmental variables. In the present study, no significant effects on sheath blight disease were detected from all other measured traits in F2:3 populations. Peng and his coworkers, 2003 concluded that for most of the morphological traits, their inheritance was independent of sheath blight resistance, and the correlations between them and sheath blight resistance were not significant or they were inconsistent in a population derived from a Jasmine85/ Lemont and by [28] in a population derived from a HP2216/Tetep. All these studies clearly showed that there are many ShB QTLs that were mapped independent of PH (plant height) and HD (heading date) on all rice chromosomes Some reports have shown correlation of sheath blight with plant height or heading date studied that sheath blight resistance as they measured it in Tetep might be due to the molecular mechanisms involved in host-pathogen interaction and not due to any morphological adaptation to avoid disease. Similarly in our study with ARC10531 we did not find any morphological correlation with disease score [12,28,37]. In our study the SSR markers RM205 and RM336 were found associated with sheath blight resistance gene/ locus in the rice cultivar ARC10531 using bulked segregant analysis. Similarly identified three major QTLs for sheath blight resistance in an F2 population of Jasmine 85/Lemont through bulked segregant analysis in their study performed the BSA using F2 population from the cross between Lemont and Teqing. The two major QTLs for rice sheath blight identified on chromosome 9 and 11 with R2 value of 12.9% and 15.3% developed an F2 population from a cross between 4011 and Xiangzaoxian19 to identify molecular markers linked with the sheath blight resistance using BSA. The dominant resistant gene/ locus named as Rsb1 was mapped on rice chromosome 5 linked with SSR marker RM164 [19] have discussed the prospects of bulk sergeant analysis in a broad range of applications in gene mapping [26,27,38]. Furthermore the genetic map of chromosome #7 and chromosome #9 was constructed to verify the existence of previous reports of QTL region linked with these two markers RM 336 and RM 205 associated with ShB resistance identified in the present study. On chromosome #7 the linked marker RM336 are in agreement with the previously reported result of [28]. Chromosome #9 of rice has been reported to contain many major effect ShB QTLs, most of which are closely mapped to each other. In the present study, SSR marker RM205 found associated with ShB resistance has been mapped earlier by [39].The consistent QTL for ShB resistance on chromosome #9 has verified by several other researchers which indicate its authenticity and stability. It once again signifies the importance of BSA in establishing marker-trait association in a rapid way. The findings of this study could be directly useful in molecular analysis of segregating generations, breeding lines and varieties having ARC10531 as a parent [10,40] (Table 5). |
| References |
|
Visit for more related articles at Rice Research: Open Access
Abstract
The population consisting of 210 F2:3 individuals from the cross between BPT-5204 (highly susceptible to sheath blight) and ARC-10531 a land race from Assam (moderately resistant to sheath blight) was analyzed to identify the markers associated with sheath blight resistance and to study any association of any morphological trait to disease incidence. The frequency distribution curve of F2:3 progenies for disease trait were continuous, indicating the polygenic control over the trait. The range of relative lesion height was 21-75% with a mean of 38.59%.No significant association between sheath blight disease and other morphological traits were detected in F2:3 populations. Parental polymorphisms were surveyed with 500 primer pairs of simple sequence repeats (SSR), revealed 70 polymorphic markers between the parents. In order to detect the major effect, QTL associated with sheath blight resistance, a strategy of combining the DNA pooling from selected segregants and genotyping was adopted. The association of putative markers identified based on DNA pooling from selected segregant was established by Single Marker Analysis (SMA).The results of SMA revealed that SSR markers, RM336 (chromosome#7) and RM205 (chromosome#9) showed significant association with sheath blight and accounted for 21.8% and 17.3% of total variation respectively. The results obtained from the DNA pooling of phenotypic extremes could be a useful strategy to detect the genetic loci with major effects of the complex trait such as disease resistance in rice.
| Keywords |
| ARC10531; Bulked segregant analysis (BSA); Rice; Sheath blight |
| Introduction |
| In the present scenario of increasing global human population, decreasing arable land, predicted increases of water scarcity, soil salinity, severe diseases, emerging resistance of pests and pathogens to pesticides and climate change pose significant challenges to modern rice research. The biotic stresses viz. , blast, stem rot, sheath blight, and bacterial blight diseases causes severe economic losses to rice productivity. Among them Sheath blight (ShB) is an important fungal disease caused by Rhizoctonia solani Kuhn causing up to 25% of yield loss and degrades rice quality. In hot and very high humid condition, yield loss can even reach as high as 50%. With the increasing application of nitrogenous fertilizers and the popularization of semi dwarf cultivars with more tillers, ShB is becoming the most serious disease in many rice-producing areas in the world [1-5]. The fungus R. solani Kuhn is soil borne pathogen which survives either as sclerotia or mycelia in plant debris. After the initial infection, the pathogen moves on the plant through surface hyphae and develops new infection structures over the entire plant, causing significant necrotic damage. The architecture of the canopy and the associated microclimate has strong effects on both the mobilization of primary inoculums and the further spread of the disease. Absolute resistance to R.solani is not available in any of the rice germplasm grown worldwide. However, it has been reported that resistance to R. solani is a typical quantitative trait controlled by polygenes in rice [6-12]. In rice because of availability of high resolution molecular maps, complete sequence information and extensive germplasm collections, mapping of quantitative trait loci (QTLs) for disease resistance such as sheath blight is feasible in crop improvement programme. In this context has reported for the first time the identification of rice QTL resistant to ShB using RFLP markers. To date, around 50 ShB resistance QTLs (ShBR QTLs) have been detected over all 12 rice chromosomes in cultivated varieties, deep-water varieties and wild species. Some of them were identified in multiple mapping populations and/or environments and not associated with either heading date (HD) or morphological traits, and they are believed to be stable ShB QTLs [9,10,12-14]. However QTL mapping is usually carried out by genotyping large number of progenies which is labor intensive, time consuming and cost-ineffective. Several strategies have been proposed to identify molecular markers near a gene/QTL of interest with reduced number of plants to be genotyped. The two main strategies are selective genotyping and bulk segregant analysis (BSA). Selective genotyping is relatively a low-cost approach to detect QTL with large effects by genotyping individuals from the two tails of the phenotypic distribution. Bulked Segregant Analysis (BSA) serves as an affordable strategy for mapping large effect QTLs by genotyping only the extreme phenotypes instead of the entire mapping population. BSA has been successfully used in rice for identifying markers linked to QTL associated with grain quality parameters blast resistance heat tolerance, drought tolerance gall midge resistance and sheath blight resistance. In the present study, bulked segregant analysis approach was used to identify large effect QTLs for sheath blight resistance and to observe the disease association with any of morphological traits [15-26]. |
| Materials and Methods |
| Plant materials |
| 40 rice germplasm lines including 8 wild, 4 land races, 26 cultivated and 2 advanced breeding lines were screened using typha bits method to identify the resistance source for sheath blight disease. A moderate level of resistance to this disease was identified in Tetep and ARC10531, a land race with the relative lesion height percentage of 21- 30%. Molecular mapping of QTLs using Tetep as a source for moderate resistance has already been carried out by several research groups. Hence ARC10531 was selected as male parent with highly susceptible elite variety BPT-5204.Crossing program and generation advancement to F2 was performed at the glass house facility, IBT, ANGRAU, Hyderabad during 2011 and 2012.The polymorphic SSR marker RM 205 and RM22565 were used to fix F1 progenies and true F1’s were forwarded to F2 generation [27,28] (Table 1). |
| DNA extraction and SSR analysis |
| Genomic DNA was isolated from frozen fresh leaf tissue of 210 F2:3 progenies along with both the parents (BPT-5204 and ARC10531) with the procedure described by [29]. The quality and quantity of DNA were estimated spectrophotometrically using a Nano Drop (ND-1000, Wilmington, USA).The final DNA concentration was adjusted to50 ng/ μl. Parental polymorphism survey involving 500 SSR markers spanning all twelve chromosomes was carried out. These SSR markers were selected based on uniform distribution across the 12 rice chromosomes. Out of the 500 SSR markers screened, seventy were found polymorphic among the parental lines (BPT- 5204 and ARC10531).The PCR reaction for SSR analysis was performed in volumes of \15 μl containing 50 ng genomic DNA, 0.2 μM each primers, 10 mM Tris-Hcl (pH 8.3), 50 mM KCl, 100 mM each of dATP, dGTP, dCTP and dTTP, 1.5 mM MgCl2 and 0.5 unit of Taq polymerase. The PCR amplification was performed on Eppendorf Mastercycler® Germany with a PCR profile of 94°C for 5 min. followed by 35 cycles of 1 min. at 94°C, 45 sec at 55°C and 1min at 72°C followed by final extension at 72°C for 10 min. The PCR products were separated on 3.5% metaphor-agarose gel and documented using a gel documentation system (BIORAD Gel Doctm XR, USA). |
| F2:3 phenotyping for sheath blight resistance and other attributes |
| The 210 individuals of F2:3 population of the cross (BPT-5204 × ARC10531) were inoculated with the local Rajendranagar isolate (AG1-IA) of Rhizoctonia solani which was obtained from Division of Plant Pathology, Directorate of Rice Research, Rajendranagar, Hyderabad-500030 in the year of 2011.The inoculums of virulent 00isolate were multiplied in Typha stem bits of 4-5 cm long soaked in typha medium (Peptone-10g, Sucrose- 20 g, K2HPO4-0.1 g, MgSO4-0.1 g dissolved in 1000 ml DDH2O pH6-6.5) [30]. Rice plants at maximum tillering stage were inoculated with R. solani by placing the typha pieces between tillers in the central region of rice hills 5-10 cm above the ground level. Rhizoctonia solani infected plants were kept in a humid chamber made of clear plastic for 2 weeks to allow disease development. Plants were grown at 28°C under 14-hrs day light in the humid chamber in the greenhouse. The humidity was maintained between 80 to 100% from the time of inoculation to disease evaluation. To evaluate sheath blight resistance or susceptibility of rice cultivars, the relative lesion height of inoculated plants were recorded 14 days after inoculation. The relative lesion height (RLH) was calculated by the following formula for scoring disease reaction: |
| Observations were recorded 14 days after inoculation and graded as per 0-9 Standard Evaluation System (SES) scale Following morphological attributes were recorded in segregating populations for a cross BPT-5204 × ARC-10531- Days to heading (days), plant height (cm), panicle length (cm), tiller no (number), effective tiller no (number), relative lesion height (percentage) [30]. |
| Marker-phenotype association analysis |
| Bulked segregant analysis (BSA) has been proposed as an efficient strategy for identifying DNA markers linked to the genes or genomic regions of interest [17]. Polymorphic markers may represent markers that are linked to a gene or QTL of interest [32]. DNA bulks of plants with extreme resistance and those with extreme susceptibility were prepared from F2:3 phenotyped progenies. This was done by pooling aliquots, containing equivalent amounts of total DNA approximately, 50 ng/μl from each of ten highly resistant and ten highly susceptible plants of the F2:3 based on phenotypic observations. 70 polymorphic SSR primer pairs were used for screening of parents and two bulk DNA samples. DNA of individual F2:3 plants that were included in bulks were also analyzed with co-segregating markers to confirm their linkage to the sheath blight disease resistance. The SSR markers found polymorphic among the parents and the bulks were used for F2:3 progeny analysis. DNA of 210 F2:3 progenies and parents were analyzed to study co- segregation of these markers. |
| Data Analysis |
| The clearly resolved amplicons of SSR were scored manually as homozygote for the allele for susceptible parent (B), homozygote for the allele for resistant parent (A) and heterozygote carrying the alleles from both parents (H) in the data sheet. Chi-square (χ2) test was performed to test the goodness of fit of the F2:3 population for the phenotyping and marker data by comparing an observed frequency distribution with an expected one. Marker-trait association was analyzed by simple linear regression method to know the association between the markers and the sheath blight score using software GenStatv14.1 Frequency distribution curve for sheath blight resistance of 210 F2:3 progenies were drawn separately using Microsoft Office 2010 Excel utility. Variability parameters were performed in F2:3 population viz. ,mean, range, skewness, kurtosis, standard deviation and simple correlation coefficients were worked out by software= GENSTAT v14.1[33]. |
| Results |
| Phenotyping of F2:3 progenies for sheath blight resistance and other morphological traits |
| The F2:3 progenies of the cross ARC10531 and BPT-5204 (210 progenies) were phenotyped for sheath blight during wet season 2012. The frequency distribution curve of F2:3 progenies for disease were continuous and near to normal distribution. The range of relative lesion height in percentage was 21-75%. In the F2:3 progenies, more individuals were distributed towards 40-50% of relative lesion height and population appears to be skewed more towards susceptible side. The mean value recorded for relative lesion height was 38.51%. The results revealed that variability for the morphological traits viz. , plant height, heading date , number of tillers, panicle length and disease score (RLH %) ranges from 70- 127, 69-33, 5-31, 5-25, 14-25 and 21- 75 respectively. The traits such as plant height, number of productive tillers and relative lesion height exhibit enough variability (Figure 1 and Table 2). |
| Rice microsatellite markers associated with sheath blight reaction using bulk segregant analysis: |
| Using the BSA method, two bulks having distinct and often contrasting phenotypes for the trait of interest are generated from a segregating population from a single cross. Seventy polymorphic markers were used for screening of parents ARC10531, BPT-5204, resistant bulk (RB), and susceptible bulk (SB) along with individuals of F2:3 populations used in respective bulks. Two markers RM 205 (chromosome#9) and RM 336 (chromosome#7) clearly distinguished susceptible bulks from resistant bulks. The F2:3 progenies were genotyped with these two primers (RM336 and RM205) to study their possible association with sheath blight resistance. Segregation pattern with marker RM336 recorded a resistant allele of donor in 43 plants, susceptible allele of recipient was amplified in 57 plants while 110 plants exhibited both the alleles (heterozygous). Similarly for marker RM205 observed 40 plants showing donor allele, 50 plants of recipient allele while remaining 120 plants depicted as heterozygotes. Genetic analysis with chi-square test indicated goodness of fit to the expected ratio of 1:2:1 for co-dominant marker indicating the association of RM336 and RM205 with sheath blight resistant gene in the present population. To determine the strength of association between the putative markers and the respective phenotypes, linear regression analysis was carried out using marker genotype as groups. The simple regression analysis between phenotypic data of sheath blight resistance and the genotypic data of SSR marker RM336 and RM 205 indicated that the marker was significantly linked with ShB resistance. The two tagged marker RM205 and RM 336 associated with ShB resistance was mapped too, using Mapmaker 3.0 (Cambridge, MA, USA) based on F2:3 genotyping data of all polymorphic markers identified on those two respective chromosomes. These results indicated the possible detection of two genetic loci for sheath blight resistance on chromosome 7 (based on map location of RM 336) and on chromosome 9 (based on map location of RM 205) (Tables 3 and 4) (Figures 2 and 3). |
| Discussion |
| Several groups have attempted to identify sources of ShB resistance by screening local accessions, cultivars, landraces, and/or advanced breeding lines. The genotypes which were most promising as sources of ShB resistance have been screened for ShB reaction in different rice growing regions [11]. Tetep is a well reported source of resistance to sheath blight and several QTLs have already been mapped by number of researchers [28,29]. The land race ARC 10531 identified in the present study was observed with equal levels of resistance as that of Tetep, used as an alternate source for ShB resistance in process of development of mapping population. The importance of land races in exploring valuable QTLs for sheath blight resistance is previously discussed by several researchers. The F2:3 progenies of the cross ARC10531 and BPT- 5204 were phenotyped for sheath blight during Kharif 2012 following standard method of screening in a hot and humid micro-chamber using the typha bits method reported by [31,34,35].The frequency distribution curve of F2:3 progenies for disease was continuous and near to normal distribution. Similar frequency distribution curve showing continuous variation involving the F2:3 progeny derived from Japonica cross was reported for ShB resistance. They also observed the curve depicting skewness towards susceptible reaction to disease [36]. Third and fourth degree statistics viz. , skewness and kurtosis of an F2:3 progenies suggests that as plant height is positively skewed (0.10) indicates that trait is associated with complementary interactions whereas heading dates (days) negatively skewed (-0.68) and associated with duplicate interaction. The remaining traits also exhibited positive skewness which is associated with complementary interactions. The skewed distribution of sheath blight trait in present study shows that it is under the control of non-additive gene action, especially epistasis and influenced by environmental variables. In the present study, no significant effects on sheath blight disease were detected from all other measured traits in F2:3 populations. Peng and his coworkers, 2003 concluded that for most of the morphological traits, their inheritance was independent of sheath blight resistance, and the correlations between them and sheath blight resistance were not significant or they were inconsistent in a population derived from a Jasmine85/ Lemont and by [28] in a population derived from a HP2216/Tetep. All these studies clearly showed that there are many ShB QTLs that were mapped independent of PH (plant height) and HD (heading date) on all rice chromosomes Some reports have shown correlation of sheath blight with plant height or heading date studied that sheath blight resistance as they measured it in Tetep might be due to the molecular mechanisms involved in host-pathogen interaction and not due to any morphological adaptation to avoid disease. Similarly in our study with ARC10531 we did not find any morphological correlation with disease score [12,28,37]. In our study the SSR markers RM205 and RM336 were found associated with sheath blight resistance gene/ locus in the rice cultivar ARC10531 using bulked segregant analysis. Similarly identified three major QTLs for sheath blight resistance in an F2 population of Jasmine 85/Lemont through bulked segregant analysis in their study performed the BSA using F2 population from the cross between Lemont and Teqing. The two major QTLs for rice sheath blight identified on chromosome 9 and 11 with R2 value of 12.9% and 15.3% developed an F2 population from a cross between 4011 and Xiangzaoxian19 to identify molecular markers linked with the sheath blight resistance using BSA. The dominant resistant gene/ locus named as Rsb1 was mapped on rice chromosome 5 linked with SSR marker RM164 [19] have discussed the prospects of bulk sergeant analysis in a broad range of applications in gene mapping [26,27,38]. Furthermore the genetic map of chromosome #7 and chromosome #9 was constructed to verify the existence of previous reports of QTL region linked with these two markers RM 336 and RM 205 associated with ShB resistance identified in the present study. On chromosome #7 the linked marker RM336 are in agreement with the previously reported result of [28]. Chromosome #9 of rice has been reported to contain many major effect ShB QTLs, most of which are closely mapped to each other. In the present study, SSR marker RM205 found associated with ShB resistance has been mapped earlier by [39].The consistent QTL for ShB resistance on chromosome #9 has verified by several other researchers which indicate its authenticity and stability. It once again signifies the importance of BSA in establishing marker-trait association in a rapid way. The findings of this study could be directly useful in molecular analysis of segregating generations, breeding lines and varieties having ARC10531 as a parent [10,40] (Table 5). |
References |
|
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 | Table 4 | Table 5 |
Figures at a glance
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 |
Relevant Topics
- Basmati Rice
- Drought Tolerence
- Golden Rice
- Leaf Diseases
- Long Grain Rice
- Par Boiled Rice
- Raw Rice
- Rice
- Rice and Aquaculture
- Rice and Nutrition
- Rice Blast
- Rice Bran
- Rice Diseases
- Rice Economics
- Rice Genome
- Rice husk
- Rice production
- Rice research
- Rice Yield
- Sticky Rice
- Stress Resistant Rice
- Unpolished Rice
- White Rice
Recommended Journals
Article Tools
Article Usage
- Total views: 16184
- [From(publication date):
November-2015 - Jul 15, 2025] - Breakdown by view type
- HTML page views : 11381
- PDF downloads : 4803
