Brief Note on Pediatric Staphylococcus Aureus Bacteremia Molecular Epidemiology
Received: 27-Jun-2022 / Manuscript No. ECR-22-69056 / Editor assigned: 30-Jun-2022 / PreQC No. ECR-22-69056 / Reviewed: 13-Jul-2022 / QC No. ECR-22-69056 / Revised: 18-Jul-2022 / Manuscript No. ECR-22-69056 / Published Date: 25-Jul-2022 DOI: 10.4172/2161-1165.1000449
Abstract
A frequent and significant human pathogen, Staphylococcus aureus has a wide range of virulence factors and a clinical spectrum of disease. In children from Australia and New Zealand, it is one of the most common causes of bacteraemia in the post-conjugate pneumococcal vaccine era. It is also the main cause of childhood skin and soft tissue infections (SSTIs), osteomyelitis, and infective endocarditis. Antimicrobial resistance to S. aureus has dramatically changed over time and by place. Methicillin-susceptible S. aureus (MSSA) continues to be the principal cause of bacteraemia, despite the fact that community and hospital clones of methicillin-resistant S. aureus (MRSA) have appeared in Australia. There are important knowledge gaps in the molecular epidemiology of MSSA and the virulence factors in children. This lack of data emphasises how crucial ongoing national molecular surveillance systems are similar to the government-funded Australian Staphylococcal Sepsis Outcome Program (ASSOP) run by the Australian Group on Antimicrobial Resistance for S. aureus bacteraemia (SAB) across the life course.
Keywords
Methicillin; leukocidin; phylogenetic; prognosis
Introduction
The impact of S. aureus toxins on the severity, management, and prognosis of SAB illness is still under investigation and needs more research. The most extensively researched S. aureus toxin, Panton- Valentine leukocidin (PVL), is controversial since it typically occurs at higher levels in non-invasive than invasive S. aureus isolates and does not seem to have an impact on outcomes for all clinical characteristics. Further paediatric-specific S. aureus molecular analyses are required to better our understanding of complicated host-pathogen dynamics, as there is few research exploring additional virulence determinants for paediatric SAB [1]. We performed a prospective genomic investigation of paediatric SAB in light of the illness burden, shifting patterns of antibiotic resistance, and ambiguities regarding the incidence and consequences of particular virulence factors. In order to understand the molecular epidemiology of SAB in children, we looked at how clinical phenotypes, severity, and outcomes were related to antibiotic resistance genes and virulence factors.
Materials and Methods
Isolate Collection
This prospective, multi-site, cross-sectional research of paediatric SAB in Australia and New Zealand included ten newborn intensive care units (NICUs), seven tertiary and secondary paediatric hospitals, and seven tertiary and level paediatric hospitals. The clinical outcomes that were looked at included 90-day all-cause mortality or a composite outcome that was predetermined to be 90-day all-cause mortality, 90- day relapse, ICU admission, or duration of stay. The procedures for the ISAIAH dataset are further discussed below and further information is provided [2]. Children under the age of 18 who presented to the study hospitals with a positive blood culture for S. aureus were prospectively enrolled. Local infectious disease or microbiology services detected episodes throughout a 24-month period (2017–2018). Also included were children with SAB who had been moved from outlying hospitals to the research locations. Blood cultures with several microbes were left out of the study [3].
Epidemiological Analyses
Laboratories used semi-automated susceptibility platforms, typical commercial blood culture methods, and bacterial identification. Each laboratory reported its own antimicrobial susceptibility data while using the CLSI M100 minimum inhibitory concentration (MIC) breakpoints. Isolates with resistant or intermediate susceptibility were labelled as non-susceptible. Isolates of multi-resistant MRSA (mrMRSA) were identified as those that were resistant to three or more non-beta-lactam antibiotic classes. For whole genome sequencing, all isolates were sent to the national AGAR S. aureus reference laboratory (WGS) [4]. On sequencing was done using 150-bp paired-end chemistry. With the use of a QubitTM 3 Fluorometer and the MagMAXTM Express-96 deep well magnetic particle processor, genomic DNA was recovered. As advised by the manufacturer, DNA libraries were created and normalised using the Nextera XT DNA Library Preparation Kit. With an estimated coverage of >60, each genome was sequenced.
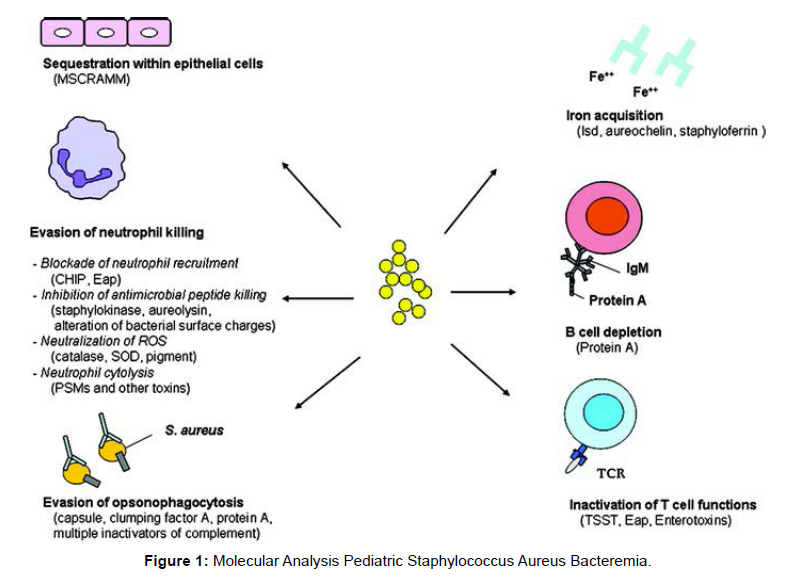
Using the SPAdes version, raw sequence reads were reconstructed from scratch. Contigs with less than 200 bp were discarded. The Supplementary Appendix S1 contains statistics on the quality of genome assembly. The assembled contigs for each isolate are accessible via a special identifier for each isolate and were uploaded to the Bacterial Isolate Genome Sequence Database (BIGSdb) on the aureus website. Genomes were annotated using the default Prokka parameters. Using Roary version, the core genes from this set were concatenated and aligned. On MEGA version X, phylogenetic trees were created using the neighbour-joining algorithm, and iTOL version 6.3 was used to visualise them [Figure 1].
By uploading each genome sequence to the PubMLST S. aureus website, MLST profiles sequence type [ST] and clonal complex [CC] were generated. The PubMLST database, where the alleles were curated, received deposits of previously unreported MLST alleles [5]. Using the SCCmecFinder online database, the staphylococcal cassette chromosome mec (SCCmec) type was identified for MRSA genomes. Following a study of the literature that looked at associations with clinical outcomes and phenotypic susceptibility, respectively, resistance and virulence genes of interest were chosen. Using BLAST, reference genes were mapped to all rebuilt genomes. The WGS data and the ISAIAH dataset were connected. The dataset included clinical data from the hospital, laboratory, and radiology records on demographics, comorbidities, infection focus, investigations, disease severity, therapy, and patient outcomes. A positive blood culture(s) collected 48 h following hospital presentation was considered evidence of community-onset SAB. The National Health Care Safety Network Centers for Disease Control and Prevention definitions of SAB linked with healthcare were modified to include SAB associated with a device or surgical site emphasis or complicating neutropenia. Repeat S [6]. aureus sterile site cultures or hospital representations that site investigators determined to be related to the initial SAB occurring 15–90 days after primary bacteraemia were considered relapses. For hospital deaths, 90-day all-cause mortality data were gathered. R version was used to perform the statistical analysis. Comparing categorical and continuous variables was done using Fisher's exact tests for categorical variables and Student's t tests or Mann-Whitney U tests for continuous variables. Covariates having a P value of 0.1 on a univariate analysis were considered potentially significant a priori and were included in the multivariable regression model. In order to prevent collinearity, pairwise correlation coefficients between variables were assessed before being added to the multivariable model. After performing stepwise backward elimination, P values were deemed statistically significant. The Hosmer-Lemeshow test and the C-statistic were both used to evaluate the performance of the model. Each laboratory hospital location provided its permission in terms of ethics.Statistical Analyses
85 percent of SAB events were caused by MSSA as the pathogen. One isolate, which the MALDI-TOF initially classified as MSSA, was later classified by the WGS as Staphylococcus argenteus. Every MRSA isolate included mecA. Only one isolate (0.2 percent) that was ST22- MRSA-IV, which was connected to the healthcare industry, was recognised as a multi-resistant MRSA, making up the majority of MRSA bacteraemia cases. Thirteen percent (38/301) of MSSA isolates were penicillin-susceptible S. aureus (PSSA); nevertheless, four of these PSSA isolates (11%) encoded a penicillinase. Between phenotypic and genotypic penicillin susceptibility testing, there was an overall 86 concordance (Supplementary Appendix S5). Cotri- moxazole resistance was found in 5% (18/353) of isolates overall, primarily in MSSA (78%) and MRSA bacteremia (22%) compared to each other.
The percentages of isolates with clindamycin inducible and constitutive resistance were 9 percent and 0.6 percent (2/353), respectively. As contrast to MRSA (10 percent, 3/31), MSSA bacteremia had a higher percentage of inducible clindamycin resistance (90 percent, 28/31); this corresponds to 84 percent concordance with genotypic testing. Results of additional antimicrobial susceptibility tests are displayed in Supplementary Appendix S5. It was shown that MSSA isolates had more STs and CCs than MRSA isolates, indicating greater variety. MRSA and MRSA isolates were genetically related, and no clusters peculiar to Australia or New Zealand were found. CCs 45, 15 and 121 were the only CCs connected to MSSA bacteraemia [7-10]. CC15 and CC8 were two popular MSSA clones related with healthcare. Eight MSSA STs were present but could not be categorised as a CC. 92 percent (24/26) of CC93 were MRSA, even though there were no CCs that contained just MRSA.
Discussion
When compared using the 90-day all-cause mortality endpoint, no changes in the frequency of any CC, virulence, or antibiotic resistance genes were found using univariate or multivariable analysis. On univariate analysis, factors for community-onset SAB included CC93 and methicillin resistance that were predictive of the composite result. However, LOS >30 days had a significant impact on this composite outcome's lone microbiological variable, PVL positive, which was the only microbiological variable included in multivariable analysis (Supplementary Appendix S6). Multifocal disease, endovascular infection, multiorgan dysfunction, surgical source control, number of surgeries, peak C-reactive protein (CRP), days with SAB and fever,LOS, and all clinical indicators of S. aureus disease severity examined on univariate analysis were all significantly associated with PVL+ SAB compared to PVL SAB. Antibacterial treatment. These findings held true for pulmonary, osteoarticular, and SSTI foci, among other SAB clinical phenotypes. S. argenteus, a coagulase-positive species in the S. aureus complex that was previously believed to be a divergent S. aureus clonal lineage, was identified as one MSSA isolate using WGS.
Conclusion
The eight-year-old from regional Australia with a communityonset single focus osteoarticular infection and bacteraemia lasting longer than the median of 1 day for the entire cohort was the source of the S. argenteus isolate found in this investigation. No relapse or death was noted after the patient had antibiotic medication for a total of five weeks. S. argenteus has been found in clinical infections in humans, non-human primates, and African bats. There is a dearth of information on the epidemiology, clinical importance, and risk of transmission of S. argenteus. New research indicates that S. argenteus's pathogenicity is comparable to that of A. aureus only one other instance of paediatric S. argenteus has been documented, and that was a Japanese child who had lymphadenitis. Our case adds significantly to the body of knowledge about the pathogenicity of S. argenteus bacteremia in paediatric settings. We further draw attention to the fact that only a small percentage of S. argenteus is found in Australian and New Zealand children who have SAB.
Acknowledgement
None
Conflict of Interest
None
References
- Liu B, Park S, Thompson CD, Li X, Lee JC (2017) Antibodies to Staphylococcus aureus capsular polysaccharides 5 and 8 perform similarly in vitro but are functionally distinct in vivo. Virulence 8(6):859-874.
- Boyle-Vavra S, Li X, Alam MT, Read TD, Sieth J, et al (2015) USA300 and USA500 clonal lineages of Staphylococcus aureus do not produce a capsular polysaccharide due to conserved mutations in the cap5 locus. mBio 6(2):585-614.
- Tuchscherr LP, Buzzola FR, Alvarez LP, Caccuri RL, Lee JC, et al (2005) Capsule-negative Staphylococcus aureus induces chronic experimental mastitis in mice. Infect Immun 73(12):7932-7937.
- O'Riordan K, Lee JC (2004) Staphylococcus aureus capsular polysaccharides. Clin Microbiol Rev 17(1):218-234.
- Visansirikul S, Kolodziej SA, Demchenko AV (2020) Staphylococcus aureus capsular polysaccharides: a structural and synthetic perspective. Org Biomol Chem 18(5):783-798.
- Echániz-Aviles G, Velazquez-Meza ME, Rodríguez-Arvizu B, Carnalla-Barajas MN, Noguerón AS (2022) Detection of capsular genotypes of methicillin-resistant Staphylococcus aureus and clonal distribution of the cap5 and cap8 genes in clinical isolates. Arch Microbiol 204(3):186.
- Wojcik-Bojek U, Rozalska B, Sadowska B (2022) Staphylococcus aureus-A Known Opponent against Host Defense Mechanisms and Vaccine Development-Do We Still Have a Chance to Win?. Int J Mol Sci 23(2):948.
- Park KH, Quaintance GKE, Cunningham SA, Chia N, Jeraldo PR, et al (2017) Molecular epidemiology of Staphylococcus aureus bacteremia in a single large Minnesota medical center in 2015 as assessed using MLST, core genome MLST and spa typing. PLoS One 12(6):179-203.
- Santosaningsih D, Santoso S, Budayanti NS, Suata K, Lestari ES, et al (2016) Characterisation of clinical Staphylococcus aureus isolates harbouring mecA or Panton-Valentine leukocidin genes from four tertiary care hospitals in Indonesia. Trop Med Int Health 21(5):610-618.
- Chiu YK, Lo WT, Wang CC (2012) Risk factors and molecular analysis of Panton-Valentine leukocidin-positive methicillin-susceptible Staphylococcus aureus colonization and infection in children. J Microbiol Immunol Infect 45(3):208-13
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Citation: C oombs S (2022) Brief Note on Pediatric Staphylococcus Aureus Bacteremia Molecular Epidemiology. Epidemiol Sci, 12: 449. DOI: 10.4172/2161-1165.1000449
Copyright: © 2022 Coombs S. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 2627
- [From(publication date): 0-2022 - Nov 24, 2025]
- Breakdown by view type
- HTML page views: 2186
- PDF downloads: 441